Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.109 n.11 Pretoria Nov. 2019
http://dx.doi.org/107196/samj.2019.v109i11.13979
RESEARCH
Intron 22 inversion real-time polymerase chain reaction detection in haemophilia A families from central South Africa
J F HoppersI, II; G Μ MarxIII; W J Janse van RensburgIV, V
IMMedSc; Department of Haematology and Cell Biology, Faculty of Health Sciences, University of the Free State, Bloemfontein, South Africa
IIMMedSc; Universitas Academic Business Unit, National Health Laboratory Service, Bloemfontein, South Africa
IIIPhD; Department of Genetics, Faculty of Natural and Agricultural Sciences, University of the Free State, Bloemfontein, South Africa
IVPhD; Department of Haematology and Cell Biology, Faculty of Health Sciences, University of the Free State, Bloemfontein, South Africa
VPhD; Human Molecular Biology Unit, School of Biomedical Sciences, Faculty of Health Sciences, University of the Free State, Bloemfontein, South Africa
ABSTRACT
BACKGROUND. Intron 22 inversion (inv22) may account for 45% of all cases of severe haemophilia A. Haemophilia A is underdiagnosed in South Africa (SA), and owing to limited resources the genotypes of most haemophilia A patients are unknown.
OBJECTIVES. To screen the haemophilia A population in central SA for inv22 using two novel detection methods.
METHODS. We recruited 62 participants from 27 families affected by haemophilia A in Free State and Northern Cape provinces. We screened for inv22 with our previously reported conventional polymerase chain reaction (PCR) method, as well as with a newly developed real-time PCR method. Sanger sequencing was performed to confirm the PCR results.
RESULTS. With the real-time PCR method, 10 of the severe haemophilia A patients and 3 carriers tested inv22-positive. The conventional PCR method and real-time PCR results were comparable in all but one case, where the discrepancy was attributed to sample-specific degradation. Inv22 was found in 29.4% of the severe haemophilia A population and 22.2% of the potential carriers. The inv22 status of most SA haemophilia A patients is currently unknown. The 29.4% of haemophilia A patients who were positive for inv22 was lower than the expected 45%, which could indicate a more prominent mutation than inv22 in the SA population.
CONCLUSIONS. The above finding needs to be confirmed by performing comprehensive factor VIII gene (F8) genotyping on the remainder of the haemophilia A patients in SA. The study contributes to genetic research in haemophilia A and lays a foundation for future research in haemophilia A genetics in SA.
Haemophilia A is an X-linked recessive bleeding disorder characterised by deficiency of coagulation factor VIII (FVIII).[1] The disorder can be classified according to procoagulant levels of FVIII: patients with severe haemophilia A have levels of < 1 IU/dL, those with moderate disease 1-5 IU/dL, and those with mild disease 5-40 IU/ dL. The disorder mainly affects males, at a rate of 1 in 5 000 - 10 000 worldwide.[1,2] The 2017 World Federation of Hemophilia survey[3] reported that 149 764 individuals were affected by haemophilia A globally, with only 1 848 reported in South Africa (SA).
Haemophilia A is underdiagnosed in SA, as in many other developing countries, owing to a lack of access to diagnostic expertise.[4] There is currently only one genetic testing facility, which services all the haemophilia A treatment centres in SA.[4] Genetic research on haemophilia is therefore limited in SA,[4] as evident from the FVIII gene (F8) variant database, to which SA has made no contribution.[5]
Genetic testing is of paramount importance in the comprehensive treatment plan for haemophilia A, since mutations in F8 are the cause of the disorder. Furthermore, F8 mutations may be associated with the development of FVIII inhibitors,[6] and variant detection allows for prenatal and carrier detection to enable genetic counselling of haemophilia A families.[7]
According to the literature, 45% of all severe haemophilia A cases are associated with an inversion in intron 22 (inv22) of f8.[8,9] Based on gene expression, an mRNA conventional inv22 polymerase chain reaction (PCR) screening method was developed to allow rapid detection of inv22.[10] This method assumes the presence of an additional exon, called exon 23c, contained in the inv22 mutant FVIII transcript.[11] Exon 23c was found to be spliced to exon 22 of F8 in unrelated haemophilia A patients.[10]
Objectives
To detect the presence of inv22 in the haemophilia A population of central SA and potential haemophilia A carriers.
Methods
Population group
Sixty-two participants were recruited from the haematology clinic at Universitas Academic Hospital in Bloemfontein and Robert Mangaliso Sobukwe Hospital in Kimberley. Twenty-seven families, represented by 34 patients with severe haemophilia A (FVIII <1 IU/dL), 3 with mild disease (FVIII 5 - 40 IU/dL), 18 potential haemophilia A carriers (mothers or sisters of known severe haemophilia A patients) and 7 healthy volunteers in Free State and Northern Cape provinces were included. The study group included all the severe haemophilia A patients and some of their family members who are actively treated at the Bloemfontein and Kimberley adult haemophilia state-operated clinics. In a recent unpublished MMed project, an internal file audit reported that these clinics had a total of 56 haemophilia A patients (M Roux, University of the Free State, unpublished data, 2018), of whom 25 with severe and 1 with mild disease were regularly followed up at a haemophilia treatment clinic. All the actively treated patients were therefore recruited for the study, as well as some of the presumed inactive patients (not regularly attending a haemophilia treatment clinic), to make a total of 37 haemophilia A patients. With a population size of just over 2 million (2 040 707) males in the Free State and Northern Cape,[12] it would be expected that the two provinces would have -204 haemophilia A patients. The relatively low number of 56 patients may be an indication of the underdiagnosed state of the disorder or, less likely, that the prevalence of the disorder is much lower in the study area than elsewhere.
Three inv22 controls were included: an inv22-negative control (CI) and 2 unrelated inv22-positive controls (C2 and C3). These controls were also receiving active treatment for haemophilia A at the Bloemfontein state-operated clinic, and had previously been screened for inv22 using the Southern blot inv22 method. In all cases, the mutation status of the other study participants and the potential carriers was unknown.
Ethics approval was obtained from the Health Sciences Research Ethics Committee of the University of the Free State (ref. no. ECUFS 166/2015) and the Free State (ref. no. FS_2015RP47_195) and Northern Cape (ref. no. NC_2015RP1_898) provincial departments of health.
RNA extraction and complementary DNA synthesis
Venous blood was collected in ethylenediaminetetra-acetic acid-containing tubes. RNA was stabilised using RNALafer solution (Ambion, Life Technologies, USA) and subsequently isolated with the RiboPure-Blood Kit (Ambion, Life Technologies, USA). A High-Capacity RNA-to-cDNA kit (Applied Biosystems, USA) was used for complementary DNA (cDNA) preparation after RNA concentrations were measured using a Qubit 2.0 Fluorometer (Invitrogen, USA).
Inv22 screening
All the participants were screened with the published conventional inv22 PCR assay[10] The Southern blot-confirmed inv22 patient samples were included in each run to serve as positive and negative inv22 controls. After gel electrophoresis, gel images were analysed to determine the inv22 genotypes of each study participant.
Additionally, a newly developed real-time PCR application was applied (Power SYBR Green Master Mix; Applied Biosystems, USA). The real-time PCR method allows for higher throughput of samples and requires less post-PCR involvement than a conventional PCR method, making it a more attractive option for high-throughput laboratories. Melt-profile analysis was performed to ensure real-time reverse transcriptase PCR reaction optimisation. Published primer pairs[9] were used and optimised for cDNA concentrations using the inv22 controls. For quality control purposes, a non-template sample and a Southern blot-confirmed positive inv22 control were included in each run, all reactions were performed in duplicate, and ROX-dye (Applied Biosystems, USA) served as internal control. Two separate real-time PCR reactions were prepared in duplicate, one containing the wild-type primers and the other the inv22 primers using the 2 χ Power SYBR Green PCR Master Mix (Applied Biosystems, USA). Cycling conditions were 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and at 62°C for 1 minute. After that, melt-curve analysis was conducted at 95°C for 1 minute and at 62°C for 1 minute, followed by 40 cycles of melting starting at 60°C with 0.5°C increments (10 seconds for each increment) of heating. The real-time assay amplification curves and melt curves were then analysed to determine the accuracy of the assay in detecting the FVIII wild-type and inv22 genotypes. The study participants were screened with the optimised real-time inv22 assay, and positive and negative controls were included in each run for quality control purposes. The amplification curves were analysed to determine the inv22 genotypes.
Sanger sequence analysis
Sanger sequencing was performed on all samples to confirm the PCR results and determine whether exon 23c is spliced to exon 22 in all positive inv22 cases.[10] Following cDNA synthesis, the amplification product of each sample was treated with ExoSAP-IT Express reagent (Affymetrix, USA). The purified PCR product consequently served as template for the sequencing reaction, using the BigDye Terminator v3.1 Kit (Applied Biosystems, USA), and impurities were removed using the ZR DNA Sequencing Clean-Up Kit (Zymo Research, USA). The sequence raw reads were analysed on the ABI Prism 3130 Genetic Analyser (Applied Biosystems, USA). The sequence of each sample was aligned (LALIGN) to the respective inv22 reference sequences. Sequence data were subsequently used to confirm the conventional and real-time inv22 PCR results.
Results
The inv22 results are summarised in Table 1. Ten (29.4%) of the haemophilia A participants were positive for inv22, 4 (22.2%) were carriers, and the rest of the participants were all inv22-negative. The inv22-positive samples displayed an expected mutant 132 base pair (bp) PCR fragment, and conversely the inv22-negative samples displayed an expected wild-type 260 bp fragment. Inv22 carriers displayed both the mutant and wild-type PCR fragments.[10]
The real-time inv22 assay detected 10 haemophilia A patients with inv22 (29.4%) and 3 inv22 carriers (16.7%). The remainder of the patients were all inv22-negative. The discrepancy in one carrier was attributed to possible sample-specific degradation, as there was a time lapse of more than a year between the conventional PCR and the real-time PCR. The same sample was used for the respective assays and was stored at -80°C. After repeated attempts, the real-time inv22 PCR failed, which could have been due to cDNA degradation. The participant could not be reached for re-sampling.
Real-time inv22 PCR results for the study participants compared well with the inv22 controls. For the real-time inv22 assay, the inv22-negative control only had amplification curves for the wild-type primer set, while the inv22-positive controls only displayed amplification curves for the mutant-type primer set. The melt-curve analysis plots for all the controls indicated the absence of primer dimer formation and multiple fragments. Fig. 1 displays the real-time inv22 PCR amplification curves for one of the haemophilia A carriers. Amplification occurred in both the wild-type and the mutant-type primer reactions, with both the inv22-positive controls and the negative control amplifying with the carrier sample.
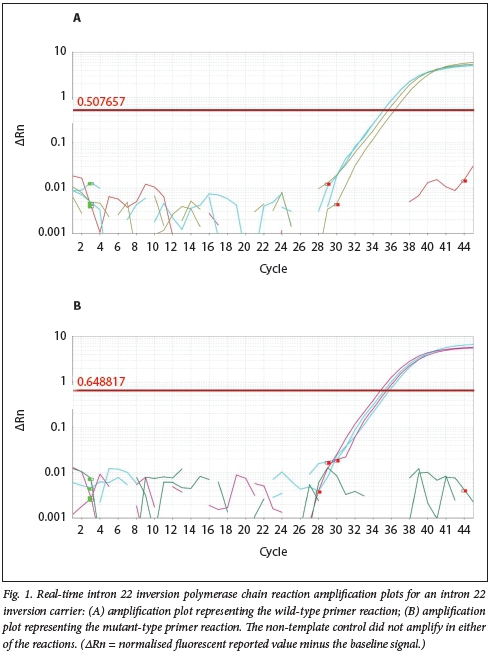
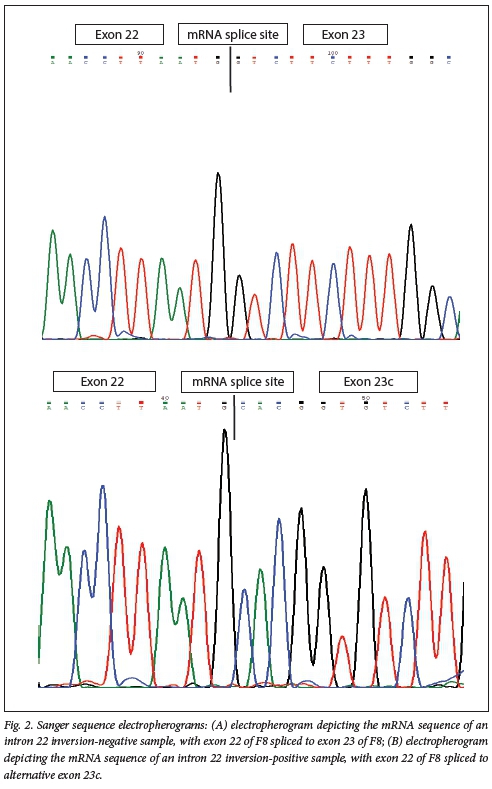
Sanger sequencing results confirmed the PCR results for both assays. Patients who were positive for inv22 had exon 23c spliced to exon 22 of F8, and inv22 carriers had both wild-type exon 22 and exon 23c associated with inv22. Fig. 2 displays the sequence electropherogram for an inv22-negative haemophilia A patient and an inv22-positive patient.
Discussion
Inv22 has been reported to affect 45% of all patients with severe haemophilia A and is the first mutation to be genotyped in cases of severe haemophilia A. We found inv22 in 29.4% of our haemophilia A study cohort in central SA, which was lower than the globally reported 45%.[8,9] The inv22-positive participants were all unrelated except for one pair of brothers, indicating that inv22 occurred in 9 of the 27 haemophilia A families (33.3%). The majority (80%) of the inv22-positive patients were black South Africans, with only 20% being white. This finding may suggest an ethnic contribution to inv22 frequencies, but will have to be confirmed with a larger study cohort. Inv22 was found in 22.2% of the potential carriers. The lower prevalence of inv22 in our study population could be significant, considering that the majority of other studies have reported an inv22 prevalence of 45%. The sample size in this study was small, however, and to confirm such a hypothesis the remainder of the SA haemophilia A population will have to be screened for inv22. Interestingly, 24% of the severe haemophilia A patients had a history of FVIII inhibitor development, and of those patients, 33% had inv22. This finding is in keeping with the prediction that between 20% and 40% of patients with FVIII inhibitors have inv22.[13,14]
In this study, we modified a rapid and cost-effective conventional inv22 PCR RNA-based screening method into a real-time inv22 PCR screening method that can be utilised by high-throughput laboratories that prefer real-time applications. The real-time inv22 PCR screening method seemed to be robust, as it compared well with the conventional inv22 PCR method and the results obtained with sequence analysis. A result discrepancy in one of the inv22 carrier participants was possibly attributed to the samples cDNA degrading during storage, as the conventional and real-time assays were done more than a year apart. We therefore recommend that the robustness of the real-time inv22 PCR screening method be confirmed with a further validation study to determine cDNA storage time stability before the assay is considered for diagnostic purposes.
Inv22 mutation status is unknown in most SA haemophilia A patients, with very few studies published on patient genotypes.[10] Possible carriers were included in the study, as carrier detection plays an important role in counselling haemophilia A families with regard to family planning and the risk associated with having a child with a bleeding disorder. In our setting, only severe haemophilia A patients actively attend our haemophilia clinics, and the population group may therefore be misrepresented with regard to mild and moderate haemophilia A patients. The literature and this study do not report mild and moderate haemophilia A patients with inv22.
Conclusions
Although a small population was investigated in this study, in our setting the haemophilia A group investigated was substantial in size considering the underdiagnosis of haemophilia A in SA,[4] and all actively treated severe haemophilia A patients in our setting were included. Future studies need to determine whether the inv22 prevalence is lower in the remainder of the SA haemophilia A population and what other causative mutations associated with haemophilia A are prevalent in this population.
F8 mutation detection has proved important in haemophilia A treatment, with specific reference to F8 genotypes that are associated with FVIII inhibitor development, [15] as well as genetic counselling.[4,6] This study lays the foundation for comprehensive genetic research to be conducted in the southern African haemophilia A population, and we hope that it will contribute to better treatment and management plans for families affected by haemophilia A on the African continent in the future.
Declaration. None.
Acknowledgements. We thank the National Research Foundation (Freestanding, Innovation and Scarce Skills Masters Scholarship awarded to JFK), the National Health Laboratory Service (Research Trust Grant awarded to WJJvR), Prof. Marius Coetzee for assistance with patient identification, and the Bloemfontein and Kimberley haemophilia outreach clinics.
Author contributions. JFK assisted in the design of the study, performed the study and wrote the manuscript. GMM assisted in the design of the study, verified the real-time PCR results and critically evaluated the manuscript. WJJvR assisted in the design of the study, supervised the study and critically evaluated the manuscript.
Funding. National Health Laboratory Service Research Trust Grant.
Conflicts of interest. None.
References
1. Hoffbrand A, Moss V Coagulation disorders. In. Essential Haematology. 6th ed. Oxford. Wiley-Blackwell, 2011.346-352. [ Links ]
2. Soucie JM, Evatt B, Jackson D. The occurrence of hemophilia in the United States. The Hemophilia Surveillance System Project Investigators. Am J Hematol 1998,59(4).288-294. https://doi.org/10.1002/(sici)1096-8652(199812)59:4<288..aid-ajh4>3.0.co,2-i [ Links ]
3. World Federation of Hemophilia. Report on the Annual Global Survey 2016. Quebec, Canada. WFH, October 2017. https://wwwl.wfh.org/publication/files/pdf-1690.pdf (accessed 30 November 2017). [ Links ]
4. Mahlangu JN, Medical and Scientific Council of the South African Haemophilia Federation. Haemophilia care in South Africa. 2004 - 2007 look back Haemophilia 2009,15(1).135-141. https://doi.org/10.1111/j.l365-2516.2008.01807.x [ Links ]
5. European Association for Haemophilia and Allied Disorders. Factor VIII gene (FS) variant database. http://www.factorviii-db.org (accessed 25 September 2019). [ Links ]
6. Jayandharan GR, Srivastava A, Srivastava A. Role of molecular genetics in hemophilia. From diagnosis to therapy Semin Thromb Hemost 2012,38(1).64-78. https://doi.org/10.1055/S-0031-1300953 [ Links ]
7. Kessler L, Adams R Mighion L, Walther S, Ganguly A. Prenatal diagnosis in haemophilia A. Experience of the genetic diagnostic laboratory. Haemophilia 2014,20(6) .e384-e391. https://doi.org/10.llll/hae.12517 [ Links ]
8. Kaufman RJ, Fay PJ, Popolo L, Ortel TL. Factor V and factor VIII. In. Marder VJ, Aird WC, Bennett JS, Schulman S, White GC, eds. Haemostasis and Thrombosis. Basic Principles and Clinical Practice. 6th ed. Philadelphia. Lippincott Wilhams & Wilkins, 2013T179-196. [ Links ]
9. Johnsen J, Fletcher S, Huston H, et al. Novel approach to genetic analysis and results in 3000 hemophilia patients enrolled in the My Life, Our Future initiative. Blood Adv 2017,1(13).824-834. https://doi.org/10.1182/bloodadvances.2016002923 [ Links ]
10. Kloppers J, Janse van Rensburg W. Rapid identification of the intron 22 inversion in haemophilia A. Hemophilia 2017,23(l).e55-e57. https://doi.org/10.1111/hae.l3142 [ Links ]
11. Pandey GS, Yanover C, Miller-Jenkins LM, et al. Endogenous factor VIII synthesis from the intron 22-inverted F8 locus may modulate the immunogenicity of replacement therapy for hemophilia A. Nat Med 2013-,19(10).1318-1324. https://doi.org/10.1038/nm.3270 [ Links ]
12. Statistics South Africa. Mid-year population estimates 2018. Statistical release P0302. Pretoria. Stats SA, 2018. https://www.statssa.gov.za/publicauons/P0302/P03022018.pdf (accessed 26 September 2019). [ Links ]
13. Astermark J, Oldenburg J, Escobar M, White II G, Berntrop E, Malmö International Brother Study group. The Malmö international Brother Study (MIBS). Genetic defects and inhibitor development in siblings with severe hemophilia A. Haematologica 200500(7):924-931. [ Links ]
14. Bardi E, Astermark J. Genetic risk factors for inhibitors in haemophilia A. Eur J Haematol 2015,94(s77).7-10. https://doi.org/10.1111/ejh.12495 [ Links ]
15. Garagiola I, Palla R Peyvandi F. Risk factors for inhibitoi development in severe hemophilia A. Thromb Res 2018,168.2027. https://doi.org/10.1016/j.thromres.2018.05.027 [ Links ]
 Correspondence:
Correspondence:
W J Janse van Rensburg
jansevrwj@ufs.ac.za
Accepted 11 April 2019














