Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
SAMJ: South African Medical Journal
versão On-line ISSN 2078-5135
versão impressa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.107 no.3 Pretoria Mar. 2017
http://dx.doi.org/10.7196/samj.2017.v107i3.12111
RESEARCH
Antibiotic resistance patterns and beta-lactamase identification in Escherichia coli isolated from young children in rural Limpopo Province, South Africa: The MAL-ED cohort
A S DeFrancescoI, II; N F TanihIII, IV; A SamieIII; R L GuerrantV; P O BessongVI
IBSc, MA; Department of Molecular and Cellular Biology, Harvard University, Cambridge, Mass., USA
IIBSc, MA; HIV/AIDS & Global Health Research Programme, Department of Microbiology, School of Mathematical and Natural Sciences, University of Venda, Thohoyandou, Limpopo, South Africa
IIIPhD; HIV/AIDS & Global Health Research Programme, Department of Microbiology, School of Mathematical and Natural Sciences, University of Venda, Thohoyandou, Limpopo, South Africa
IVPhD; Present address: Medical Research Council Unit, Fajara, The Gambia
VMD; Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, Va., USA
VIPhD, MSc; HIV/AIDS & Global Health Research Programme, Department of Microbiology, School of Mathematical and Natural Sciences, University of Venda, Thohoyandou, Limpopo, South Africa
ABSTRACT
BACKGROUND. Antibiotic resistance is a growing problem worldwide. Mechanisms of resistance vary, and some can confer resistance to multiple classes of antibiotics.
OBJECTIVE. To characterise the antibiotic resistance profiles of Escherichia coli isolates obtained from stool samples of young rural children exposed or unexposed to antibiotics.
METHODOLOGY. The samples were collected from children aged 4 - 12 months who were participants in the Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) project at the South Africa research site. We isolated 87 E. coli samples (clones) from 65 individual participants, all of which were subjected to disc diffusion assay to determine resistance. We characterised the minimum inhibitory concentration of antibiotics in a subset of strains as well as the mechanism by which these strains were resistant to beta-lactam antibiotics.
RESULTS. Our results revealed high resistance rates to co-trimoxazole (54.0%), penicillin (47.1%) and tetracycline (44.8%) in our isolates, and indicated that the beta-lactamase TEM-1 is a prevalent source of beta-lactam resistance. We also identified two isolates with the extended-spectrum beta-lactamase CTX-M-14.
CONCLUSIONS. This study identified antibiotic-resistant E. coli in children with and without prior exposure to antibiotics, with some isolates showing resistance to multiple classes of antibiotics. Clinicians should bear in mind that transmission of extended-spectrum beta-lactamase-resistant E. coli exists at the community level, and that children as young as 2 years may be harbouring these resistant phenotypes.
The problem of antibiotic resistance is not a new one. Multiple drug resistance in Escherichia coli was first observed in the 1950s. Transfer of resistance between species had also been observed by this time.[1] There has been evidence for the transfer of resistance genes between members of the human microbiota, as well as from livestock-associated bacteria to human-associated bacteria.[2] The presence of antimicrobial resistance genes even in non-pathogenic isolates therefore represents a problem, as these genes can easily be transferred to a pathogen.
Children acquire bacteria from their mother during birth,[3,4] and their gut microbiomes then undergo maturation during the first 3 years of life.[5] The early colonisation and development of this dynamic environment may predispose individuals to differences in disease incidence and outcomes. Considering that intestinal infectious diseases are the leading cause of death in children aged <14 years in Limpopo Province, South Africa (SA),[6] and that resistance genes can be geographically distinct, identification and monitoring of resistance mechanisms is important in order to foster appropriate treatment regimens. We therefore decided to focus on community isolates from children as opposed to clinical isolates, which are often the source of strains in studies focused on resistant organisms.
The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) project was designed to look for correlations between factors present during childhood in developing regions, focusing on the relationship between enteric pathogen presence and growth and development outcomes. Each participant in MAL-ED had stool samples taken on at least a monthly basis from birth until age 2 years, and information on health events such as the incidence of diarrhoea and exposure to antibiotics was collected. In addition, developmental milestone data for each participant were recorded, such as height, weight and cognitive ability. This rich data set allowed us to consider which antibiotics were most relevant to the community, as we had details of exposure for all the study participants (supplementary Table 1: Appendix 1). Penicillin-class antibiotics are the most frequently administered, so resistance to this class would be most detrimental to health outcomes; we therefore determined that beta-lactamase genes would be an appropriate area of focus.
Objective
To present data on E. coli strains isolated from stool samples collected as part of the MAL-ED study. The children from whom the strains were isolated ranged in age from 4 to 12 months and had varying histories in terms of antibiotic exposure and diarrhoeal events. The antibiotic susceptibilities of the isolates were characterised, along with their minimum inhibitory concentrations (MICs) and identification of some of the beta-lactamase genes responsible. For beta-lactam-resistant isolates, we tested for variants of several narrow-spectrum beta-lactamases (TEM, SHV and OXA), as well as some variants of the CTX-M extended-spectrum beta-lactamase (ESBL) type.
Methods
Ethical considerations
The study protocol was approved by the Research Ethics Committee of the University of Venda, SA (ref. no. SMNS/09/MBY/004). Permission was obtained from the Department of Health, Limpopo Province (ref. no. 4/2/2), SA. Signed informed consent was obtained from the parents or legal guardians of all study subjects prior to enrolment and sample collection.
Strain isolation and growth
E. coli strains were isolated from stool samples collected as part of the MAL-ED study, and in addition E. coli ATCC 25922 (Microbiologics, USA) was maintained as a control strain. Initial isolation of lactose fermenters (pink colonies) was performed on MacConkey agar (Neogen, USA), followed by screening on EMB agar (Neogen, USA), on which E. coli produce a characteristic green sheen. In total, 87 strains were isolated from 65 study participants. Cultures were maintained in nutrient broth (Oxoid, UK) or on nutrient agar (Neogen, USA) at 37°C.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed using the disc diffusion assay.[7] Briefly, colonies were resuspended in sterile saline to a McFarland standard of 0.5, and were uniformly spread on Mueller-Hinton agar (Mast Diagnostics, UK) using sterile cotton swabs. Antibiotic discs (Mast Diagnostics, UK) were dispensed using the Discmaster 3 Dispenser (Mast Diagnostics, UK), and plates were incubated overnight at 37°C. Zones of inhibition were then measured against a black, non-reflecting background. E. coli ATCC 25922 was utilised as a control strain to ensure that zones of inhibition were within an appropriate range, according to Clinical and Laboratory Standards Institute (CLSI) standards.[7] The CLSI zone diameter interpretive criteria for Enterobacteriaceae were used for interpretation of all antibiotic inhibitory zones except those of the macrolides. For azithromycin, a zone <13 mm was considered to indicate resistance, a zone of 14 - 17 mm was considered the intermediate range, and a zone >18 mm was considered to indicate sensitivity.
Detection and identification of beta-lactamases
Multiplex polymerase chain reactions (PCRs) for detection of beta-lactamase genes were designed by Dallenne et al.[8] The first multiplex PCR amplified blaTEM/blaSHV/blaOXA-1-like genes using the following primers: MultiTSO-T_for CATTTCCGTGTCGCCCTTATTC (0.4 μM)/MultiTSO-T_rev CGTTCATCCATAGTTGCCTGAC (0.4 μM) - product size 800 bp, MultiTSO-S_for AGCCGCTTGAGCAAATTAAAC (0.4 μM)/MultiTSO-S_rev ATCCCGCAGATAAATCACCAC (0.4 μM) - product size 713 bp, and MultiTSO-O_for GGCACCAGATTCAACTTTCAAG (0.4 μM)/ MultiTSO-O_rev GACCCCAAGTTTCCTGTAAGTG (0.4 μM) -product size 564 bp. The second amplified blaCTX-M phylogenetic groups 1, 2 and 9 using the following primers: MultiCTXMGp1_for TTAGGAARTGTGCCGCTGYA (0.4 μM)/MultiCTXMGp1-2_rev CGATATCGTTGGTGGTRCCAT (0.2 μM) - product size 688 bp, MultiCTXMGp2_for CGTTAACGGCACGATGAC (0.2 μM)/MultiCTXMGp1-2_rev CGATATCGTTGGTGGTRCCAT (0.2 μM) - product size 404 bp, and MultiCTXMGp9_for TCAAGCCTGCCGATCTGGT (0.4 μM)/MultiCTXMGp9_rev TGATTCTCGCCGCTGAAG (0.4 μM) - product size 561 bp. PCRs were performed in duplicate, one replicate using DNA isolated with the GeneJET Plasmid Miniprep Kit (Thermo Scientific, USA) and one using colonies resuspended in 100 μL of water and subjected to heating at 95°C for 10 minutes.
PCRs were performed using DreamTaq Green Master Mix (Thermo Scientific, USA) in 50 μL reactions, using the primer concentrations specified above. The amplification reaction was performed as in Dallenne et al.:[8] 94°C for 10 minutes; [94°C for 40 seconds, 60°C for 40 seconds and 72°C for 1 minute] χ 30 cycles; 72°C for 7 minutes. Amplified beta-lactamase genes were visualised under ultraviolet light following separation on a 2% agarose gel containing ethidium bromide, along with a GeneRuler 100 bp Plus ladder (Thermo Scientific, USA). Products were purified using the GeneJET PCR Purification Kit (Thermo Scientific, USA) and sent for sequencing (Inqaba Biotec, SA).
Sequencing results were analysed using Geneious 7 (http://www.geneious.com), and the data were searched using the NCBI Nucleotide BLAST Megablast tool (http://blast.ncbi.nlm.nih.gov/), optimised for highly similar sequences.
Minimum inhibitory concentrations
MICs were determined using MIC Test Strips (Liofilchem, Italy). Antimicrobial susceptibility testing was performed according to the manufacturer's guidelines. Briefly, colonies of each strain were suspended in sterile saline solution to achieve a 0.5 McFarland standard turbidity level, and were uniformly spread on Mueller-Hinton agar (Mast Diagnostics, UK) using sterile cotton swabs. Once the agar surface was completely dry, an MIC test strip was applied to each plate with sterile forceps and the plates were incubated at 37°C for 18 - 24 hours. The MIC was read where inhibition of growth intersected the strip.
Results
Antibiotic susceptibility of isolates
Each E. coli clone was tested for susceptibility to 13 antibiotics or combination therapies using the disc diffusion method.[7] Results of testing are shown in Table 1. The highest incidence of resistance was to the trimethoprim-sulfamethoxazole combination antibiotic (co-trimoxazole), with 54.0% of isolates showing resistance. The second highest incidence of resistance was towards penicillin-class antibiotics, with 47.1% of isolates showing resistance to ampicillin and amoxicillin. Much of this resistance appeared to be reversed by the inclusion of a beta-lactamase inhibitor, clavulanic acid. We did not observe resistance to several antibiotics including imipenem, ciprofloxacin, and both aminoglycosides tested. Although not typically included in Gram-negative susceptibility studies owing to their inefficient penetration of the cell wall,[9] a macrolide antibiotic was tested, as E. coli has been shown to be a potential reservoir for macrolide resistance genes.[10] The azithromycin results are reported in Table 1, and indicate that ~20% of isolates show some enhanced resistance to this class.
Isolates showing no resistance were most abundant (n=29), whereas the second-largest subset of isolates (n=24) showed resistance to three antibiotics, indicating that resistance to more than one antibiotic is more common than resistance to one (n=14 isolates) or two (n=7). Additional information detailing which resistances were observed in which clones can be found in supplementary Table 2 (Appendix 2).
Presence and identity of beta-lactamase genes
All strains resistant to penicillins were subjected to multiplex PCR amplification to determine which beta-lactamases were present. The multiplex PCRs, originally designed by Dallenne et al.,[8] can identify the presence of certain TEM, SHV, OXA and CTX-M variants. TEM, SHV and OXA are narrow-spectrum beta-lactamases, whereas CTX-M is an ESBL. The 41 strains tested included one isolate with resistance to the combination treatment of amoxicillin + clavulanic acid. Results of multiplex PCR are shown in Table 2. A subset of the PCR products were sequenced and analysed by BLAST, which revealed that both of the CTX-M group 9 beta-lactamases present were in fact CTX-M-14. As for the samples positive for TEM, products from 18 clones were sent for sequencing, which revealed that all 18 were TEM-1. This result is not surprising considering that this is the most frequently observed resistance gene in enterobacteria.[11] The sequences of the amplified regions can be found in supplementary Table 3 (Appendix 3).

Minimum inhibitory concentrations
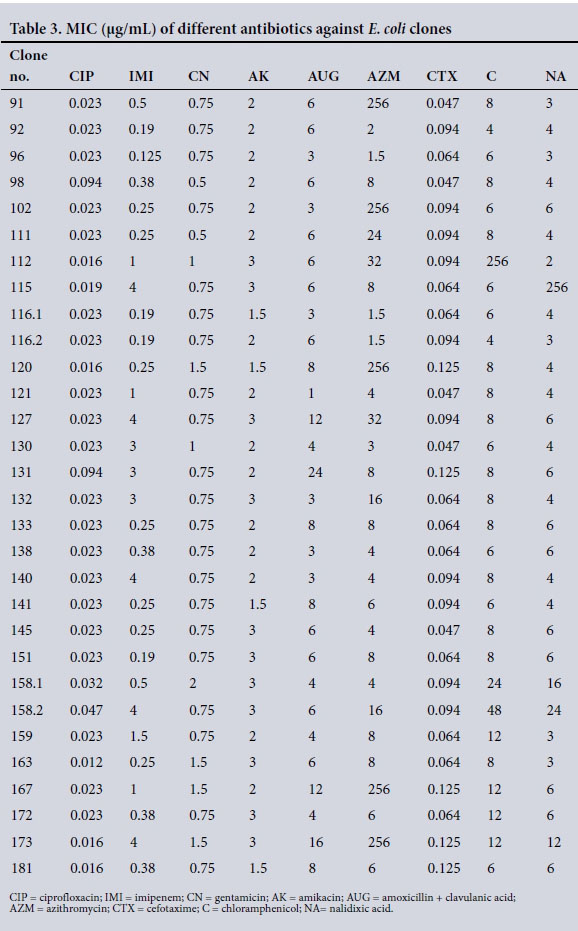
MIC ranges were 0.016 - 0.094 μg/mL for ciprofloxacin, 0.125 -4.0 μg/mL for imipenem, 0.38 - 2 μg/mL for gentamicin, 1.5 - 3 μg/ mL for amikacin, 1.0 - 24.0 μg/mL for amoxicillin + clavulanic acid, 1.5 - 32 μg/mL for azithromycin, 0.047 - 0.125 μg/mL for cefotaxime, 4.0 - 48 μg/mL for chloramphenicol and 0.125 - 24.0 μg/ mL for nalidixic acid (Table 3). Some MIC values outside the test range were observed for a subset of strains tested when exposed to chloramphenicol and nalidixic acid.
Strains showing resistance phenotypes to beta-lactam antibiotics were subjected to multiplex PCR to determine the presence of specific beta-lactamase genes. A subset of these products was sequenced, allowing identification of the specific beta-lactamase gene using BLAST.
Discussion and conclusion
When deciding which antibiotics to test in the disc diffusion assays, we considered the most frequently used antibiotics in the SA site MAL-ED participant group (supplementary Table 1). This provided useful information on the local clinical usage of various classes of antibiotics, with the penicillin class being most common, followed by sulfonamides, macrolides and others. With penicillin-class antibiotics being most commonly employed in treatment of illness, resistance to this class would have the most negative impact, so we chose to focus on resistance mechanisms to this class. Penicillin resistance was in fact the second most prevalent in our study, after co-trimoxazole resistance, which is a worrying trend considering the common use of penicillin in the management of bacterial infections.
Multidrug resistance is resistance to multiple classes of antibiotics. Of the 87 isolates tested, 2 showed resistance to five antibiotics (ampicillin/amoxicillin, co-trimoxazole, chloramphenicol, tetra-cycline and nalidixic acid) and 11 showed resistance to four antibiotics. Interestingly, the two most multidrug-resistant isolates were from individuals with no reported exposure to antibiotics. The two strains that harbour the CTX-M-14 resistance gene both show resistance to three antibiotics: cefotaxime, ampicillin/amoxicillin and tetracycline. Although antibiotic exposure increases the selective pressure for organisms to develop and maintain antibiotic-resistant elements, the presence of resistance genes apparently does not necessarily correlate with prior exposure to antimicrobial agents.
To characterise the range of MICs for our isolates, a subset was tested using MIC test strips. Results for amikacin, amoxicillin + clavulanic acid, chloramphenicol, cipro-floxacin, cefotaxime, gentamicin and nalidixic acid were all consistent with the disc diffusion assay results. For imipenem, ~16% of the MIC results indicated resistance where the disc diffusion assay had not indicated this, as did ~7% for azithromycin. This indicates that the proportion of resistant isolates may actually be higher than we reported. There was also one azithromycin test that showed resistance in the disc diffusion and sensitivity in the MIC testing.
Some participants in the MAL-ED study went on to develop severe acute malnutrition. Treatment for this condition often involves administration of antibiotics, as recommended by the World Health Organization (WHO); however, there is no strong evidence that this is the best course of action.[12] Considering our evidence that even without antibiotic exposure, children at risk of malnutrition often harbour resistance genes, it seems that the introduction of additional selective pressure could actually contribute to ill health rather than recovery. A 2014 study[13] found that the diversity of antibiotic resistance genes in the human gut microbiota appears to increase with age, although they did not look at individuals <3 years of age. This would indicate that even without antibiotic administration, the burden of resistance would increase over time.
The diversity of beta-lactamases is high, and many novel variants are reported year after year. In the early 1990s there were fewer than 150 known beta-lactamases, and by 2009, over 890 unique beta-lactamase sequences had been identified.[14] New enzymes often emerge in isolated areas and go on to expand their host range and also their geographical range. TEM-1, which we observed in a majority of penicillin-resistant isolates, is the most commonly found secondary beta-lactamase in ampi-cillin-resistant E. coli,[11]with much greater prevalence than TEM-2, SHV and OXA-1 (the other narrow-spectrum beta-lactamases that we tested for).[15] ESBLs are beta-lactamases with enhanced activity against cephalosporins, early examples of which were similar to TEM and SHV. The CTX-M-type ESBL was first observed in the late 1980s, and is not TEM- or SHV-derived.[16]
Although previous studies in SA found examples of multiple ESBLs being produced by clinical isolates,[17-20] the first report of CTX-M-type ESBLs was not until 2003, where CTX-M-2 and CTX-M-3 were found in Klebsiella pneumoniae21Since then, other CTX-M types have been found in SA, including CTX-M-14, CTX-M-15 and CTX-M-37.[22-25] The WHO reports that in the African Region there are insufficient data concerning antibiotic resistance,[26] so additional reports such as this are important in this regard. The data presented are limited to the MAL-ED SA Dzimauli community study site of Limpopo Province.[27] However, our identification of the CTX-M-14 ESBL in community E. coli isolates, even in young children who have not received antibiotics, adds to the greater picture of the antibiotic resistance landscape in SA and in the African Region.
The prescribing patterns of antibiotics, either through excessive use or through sub-therapeutic doses, in addition to the use of antibiotics in animal production, are factors known to contribute to the development and spread of antibiotic resistance at community level. Our finding has clinical relevance. Firstly, it adds to the body of evidence on the spread of antibiotic resistance in rural communities, and secondly, it supports the increasing need for a reduction in the frequency of empiricaly prescribing antibiotics, a common practice in communities without diagnostic laboratory support. Discouraging empirical prescription has been proposed in the SA national approach to 'antibiotic stewardship'.[28] In conclusion, clinicians and public health practitioners should bear in mind that transmission of ESBL-resistant E. coli exists at the community level and that children as young as 2 years, even without prior exposure to antibiotics, may be harbouring these resistant phenotypes, an awareness that should guide prescription practices.
Acknowledgements. The authors thank the staff and participants of the MAL-ED network for their important contributions. MAL-ED is carried out as a collaborative project supported by the Bill & Melinda Gates Foundation, the Foundation for the National Institutes of Health and the National Institutes of Health, Fogarty International Center. Funding for ASD was provided by the NSF GROW with USAID programme. NFT was supported by award no. D43 TW009359 from the Fogarty International Center/National Institutes of Health.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Foundation for the National Institutes of Health or the Bill & Melinda Gates Foundation. Study sponsors were not involved in the study design, collection, analysis or interpretation of data, writing of the manuscript, or the decision to submit the manuscript for publication.
Author contributions. POB, RLG and ASD conceived and designed the study, ASD and NFT carried out laboratory analysis, and ASD analysed the data and prepared the first draft. All authors made significant intellectual contributions in finalising the manuscript, and read and approved the final version for submission.
REFERENCES
1. Watanabe T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev 1963;27(1):87-115. http://dx.doi.org/10.1101/sqb.1953.018.01.037 [ Links ]
2. Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011;480(7376):241-244. http://dx.doi.org/10.1038/nature10571 [ Links ]
3. Schultz M, Gottl C, Young RJ, Iwen P, Vanderhoof JA. Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J Pediatr Gastroenterol Nutr 2004;38(3):293-297. http://dx.doi.org/10.1097/00005176-200403000-00012 [ Links ]
4. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107(26):11971-11975. http://dx.doi.org/10.1073/pnas.1002601107 [ Links ]
5. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486(7402):222-227. http://dx.doi.org/10.1038/nature11053 [ Links ]
6. Statistics South Africa. Mortality and Causes of Death in South Africa, 2013: Findings from Death Notification. Pretoria: Statistics South Africa, 2014. [ Links ]
7. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 22nd Informational Supplement. CLSI document M100-S22. Wayne, Penn.: CLSI, 2012. [ Links ]
8. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010;65(3):490-495. http://dx.doi.org/10.1093/jac/dkp498 [ Links ]
9. Vaara M. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in Gram-negative enteric bacteria. Antimicrob Agents Chemother 1993;37(2):354-356. http://dx.doi.org/10.1128/AAC.37.2.354 [ Links ]
10. Phuc Nguyen MC, Woerther PL, Bouvet M, Andremont A, Leclercq R, Canu A. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis 2009;15(10):1648-1650. http://dx.doi.org/10.3201/eid1510.090696 [ Links ]
11. Livermore DM. Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev 1995;8(4):557-584. [ Links ]
12. Alcoba G, Kerac M, Breysse S, et al. Do children with uncomplicated severe acute malnutrition need antibiotics? A systematic review and meta-analysis. PLoS One 2013;8:e53184. http://dx.doi.org/10.1371/journal.pone.0053184 [ Links ]
13. Lu N, Hu Y, Zhu L, et al. DNA microarray analysis reveals that antibiotic resistance-gene diversity in human gut microbiota is age related. Sci Rep 2014;4:4302. http://dx.doi.org/10.1038/srep04302 [ Links ]
14. Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 2010;54(3):969-976. http://dx.doi.org/10.1128/AAC.01009-09 [ Links ]
15. Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: A clinical update. Clin Microbiol Rev 2005;18(4):657-686. http://dx.doi.org/10.1128/CMR.18.4.657-686.2005 [ Links ]
16. Bonnet R. Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob Agents Chemother 2004;48(1):1-14. http://dx.doi.org/10.1128/AAC.48.L1-14.2004 [ Links ]
17. Pitout JD, Thomson KS, Hanson ND, Ehrhardt AF, Moland ES, Sanders CC. Beta-lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother 1998;42(6):1350-1354. [ Links ]
18. Hanson ND, Smith Moland E, Pitout JD. Enzymatic characterization of TEM-63, a TEM-type extended spectrum beta-lactamase expressed in three different genera of Enterobacteriaceae from South Africa. Diagn Microbiol Infect Dis 2001;40(4):199-201. http://dx.doi.org/10.1016/S0732-8893(01)00266-8 [ Links ]
19. Essack SY, Hall LM, Pillay DG, McFadyen ML, Livermore DM. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum beta-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob Agents Chemother 2001;45(1):88-95. http://dx.doi.org/10.1128/AAC.45.1.88-95.2001 [ Links ]
20. Pitout JDD, Reisbig MD, Venter EC, Church DL, Hanson ND. Modification of the double-disk test for detection of Enterobacteriaceae producing extended-spectrum and AmpC-lactamases. J Clin Microbiol 2003;41(8):3933-3935. http://dx.doi.org/10.1128/JCM.4L8.3933-3935.2003 [ Links ]
21. Paterson DL, Hujer KM, Hujer AM, et al. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob Agents Chemother 2003;47(11):3554-3560. http://dx.doi.org/10.1128/AAC.47.11.3554-3560.2003 [ Links ]
22. Segal H, Elisha BG. Resistance to β-lactams, and reduced susceptibility to carbapenems, in clinical isolates of Klebsiella pneumoniae due to interplay between CTX-M-15 and altered outer membrane permeability. South Afr J Epidemiol Infect 2006;21(2):41-44. http://dx.doi.org/10.1080/10158782.2006.11441262 [ Links ]
23. Govinden U, Mocktar C, Moodley P, Sturm AW, Essack SY. CTX-M-37 in Salmonella enterica serotype Isangi from Durban, South Africa. Int J Antimicrob Agents 2006;28(4):288-291. http://dx.doi.org/10.1016/j.ijantimicag.2006.05.028 [ Links ]
24. Usha G, Chunderika M, Prashini M, Willem SA, Yusuf ES. Characterization of extended-spectrum beta-lactamases in Salmonella spp. at a tertiary hospital in Durban, South Africa. Diagn Microbiol Infect Dis 2008;62(1):86-91. http://dx.doi.org/10.1016/j.diagmicrobio.2008.04.014 [ Links ]
25. Peirano G, van Greune CH, Pitout JD. Characteristics of infections caused by extended-spectrum beta- lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn Microbiol Infect Dis 2011;69(4):449-453. http://dx.doi.org/10.1016/j.diagmicrobio.2010.11.011 [ Links ]
26. World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. Geneva: WHO, 2014. [ Links ]
27. Bessong PO, Nyathi E, Mahopo TC, Netshandama V; MAL-ED South Africa. Development of the Dzimauli community in Vhembe District, Limpopo province of South Africa, for the MAL-ED cohort study. Clin Infect Dis 2014;59(Suppl 4):S317-S324. http://dx.doi.org/10.1093/cid/ciu418 [ Links ]
28. Goff DA, Kullar R, Goldstein EJ, et al. A global call from five countries to collaborate in antibiotic stewardship: United we succeed, divided we might fail. Lancet Infect Dis 2016;17(2):e56-e63. http://dx.doi.org/10.1016/S1473-3099(16)30386-3 [ Links ]
 Correspondence:
Correspondence:
P O Bessong
bessong@univen.ac.za
Accepted 10 January 2017

Supplementary Table 1- Click to enlarge

Supplementary Table 2 - Click to enlarge















