Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.106 n.12 Pretoria Dec. 2016
http://dx.doi.org/10.7196/SAMJ.2016.V106I12.10769
RESEARCH
Screening for calreticulin mutations in a cohort of patients suspected of having a myeloproliferative neoplasm
A de KockI; C BooysenII
IPhD; Haematology Department, Tissue Typing Laboratory, Universitas Academic Hospital, Bloemfontein, South Africa
IIBMedSc Hons Haematology Department, Tissue Typing Laboratory, Universitas Academic Hospital, Bloemfontein, South Africa
ABSTRACT
BACKGROUND. The discovery of calreticulin (CALR) has shown it to be the second most frequent mutation after the Janus Kinase 2 (JAK2) mutation in myeloproliferative neoplasms (MPNs). Its structure indicates various functions, of which two are to ensure calcium homeostasis and proper folding of other target proteins. Over 36 types of CALR mutations have been identified, all causing a recurrent frameshift in the C-terminal domain affecting CALR's localisation and calcium-binding function.
OBJECTIVE. To screen a cohort of 89 patients suspected of having an MPN for the CALR mutations.
METHODS. Capillary and gel electrophoresis were used in conjunction as confirmatory tests to screen the cohort of patients.
RESULTS. Of three samples containing a type 1 CALR mutation, two were heterozygous and one homozygous for a 52-base pair deletion in CALR.
CONCLUSIONS. Most studies report CALR mutations to be present only in patients with primary myelofibrosis or essential thrombocythaemia, with mutual exclusivity to JAK2 mutations. The findings of this study indicate that JAK2 and CALR mutations are no longer considered mutually exclusive. Similarly, patients with a polycythaemia vera phenotype could also carry a CALR mutation.
Myeloproliferative neoplasms (MPNs) can be classified according to the 2008 World Health Organization classification[1] into Philadelphia (Ph) chromosome-negative and positive neoplasms. Ph chromosome-negative neoplasms include polycythaemia vera (PV), essential thrombocythaemia (ET) and primary myelofibrosis (PMF), whereas chronic myeloid leukaemia (CML) comprises Ph chromosome- positive neoplasms.
The Janus Kinase 2 (JAK2) mutations found in 50 - 60% of patients encode a cytoplasmic tyrosine kinase protein active in the JAK/signal transducer and activator of transcription proteins (STAT) signalling pathway. A gain-of-function mutation replaces a guanidine base with a thymine base, resulting in a valine-to-phenylalanine alteration,[2] and consequently constitutive tyrosine kinase activity along with enhanced proliferation of myeloid cells. In 5 - 10% of patients the thrombopoietin receptor (MPL) gene is impaired, while in 30 - 45% of patients no specific molecular marker had been described until the discovery of calreticulin (CALR). In these 30 - 45% of patients CALR was shown to be the second most frequent mutation after JAK2, offering mutual exclusivity to JAK2 and MPL mutations, and being absent in PV patients.[2]
The CALR gene, mapped to chromosome 19p,[3] has three structural and functional domains, referred to as the N, P and C domains.'41 The N domain is important for proper folding of CALR and enables its interaction with numerous other protein molecules such as α-integrin. The P domain comprises specific amino acid sequence repeats permitting binding of high-affinity calcium ions (Ca2+) and lectin-like chaperoning.[5] The C domain contains a KDEL signal comprising four acidic amino acids (K = Lys, D = Asp, E = Glu, L = Leu) that allow CALR to have a high capacity for binding Ca2+, thus regulating CALR's interaction with other protein molecules.[6]
Of the 36 types of insertions and deletions identified, type 1 (a 52-base pair deletion) and type 2 (a 5-base pair insertion) mutations account for >80% of CALR mutations.[7] Phenotypic differences between type 1 and type 2 carriers have been implicated.'31 All recurrent mutations cause a frameshift in the region encoding the C domain, leading to altered subcellular localisation'21 and impaired function. Two main functions have been indicated, a chaperone function essential in protein processing and transport in the endoplasmic reticulum (ER), ultimately eliminating incorrectly folded proteins, and a regulatory function over intracellular Ca2+ levels whereby CALR acts as a sensor and buffer to Ca2+ in the ER lumen. Despite inconclusive evidence regarding the mechanism by which CALR assists protein folding and Ca2+ homoeostasis,[5] its depletion has profound effects.
Methods
Samples
Eighty-nine archived samples from patient and control groups, including all samples of patients suspected of having an MPN, were screened. There were JAK2-positive and negative and ET, PV and PMF samples. The patient group consisted of JAK2-negative samples, whereas the control group consisted of JAK2-positive samples, as CALR and JAK2 are thought to be mutually exclusive. DNA was extracted and purified using the Promega Purification Kit (Promega, USA) (www.promega.com/protocols/).
Polymerase chain reaction (PCR) and amplicon analysis
Published primers[2] targeting exon 9 of CALR were used (Applied Biosystems, UK). A primer-based sizing assay was used to detect CALR mutations, the forward primer (GGCAAGGCCCTGAGGTGT) being labelled with 6-carboxyflourescein (6-FAM). The reverse primer sequence was GGCCTCAGTCCAGCCCTG.
Hot-start PCR was used. The amplicons labelled with 6-FAM were electrophoresed in a 3130 Genetic Analyser (Applied Biosystems, UK), as well as on agarose gel.
A mixture of 10 μL Hi-Di Formamide and 0.5 μL GeneScan LIZ 500 (size standard), both from Life Technologies, USA, was prepared for capillary electrophoresis, of which 10 μL was added to 1.5 μL. PCR product. Nested PCR was performed for the samples subjected to gel electrophoresis, whereby the PCR products were amplified for a second time in an ABI 2700 thermocycler (Applied Biosystems, USA). PCR products were separated on a 2.5% agarose gel and analysed under ultraviolet light. Samples positive for a mutation were band-stabbed and placed in TE-1 buffer (EDTA 0.5M, Tris 1M) overnight at 4°C. Four μL of the supernatant was used for sequencing.
The wild-type allele has 267 base pairs.[8] Any fragments that differed in size to the wild-type allele carried a mutation.
DNA sequence analysis
Sequencing of samples was performed using the BigDye Terminator v3.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, USA). The sequencing products were purified using the ethanol/sodium acetate precipitation method. A volume of 25 μΐ Hi-Di Formamide was added to the pellet and the samples were run on an ABI Prism 3130 Genetic Analyser (Applied Biosystems, UK).
The sequencing reactions were analysed using Sequencing Analysis Software (version 5.3.1) (Applied Biosystems, UK). Two free online programs were used, Chromas version 2.4.3 (Technelysium, Australia) (www.technelysium.com.au/chromas_lite. html), which visualised the sequencing data, and Lalign (www.ch.embnet.org/software/ LALIGN_form.html), which compared the sequencing data with a reference sequence obtained from Ensembl[8] using the algorithm of Huang and Miller.[9]
Ethical considerations
The University of the Free State Faculty of Health Sciences Ethics Committee granted ethical approval for this study (ref. no. ETOVS 15/8).
Results
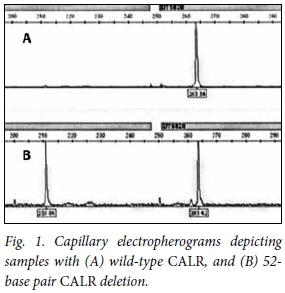
The melting temperatures for all reactions were optimised at 69°C. Of the 89 samples, only three showed a 52-base pair deletion. With two samples, two distinct peaks were observed, one at 263 base pairs and the other at 210 base pairs. With the other sample, one distinct peak was observed at 210 base pairs, making it homozygous for the type 1 deletion. Fig. 1 depicts electropherograms of (A) a patient with no CALR mutation present, and (B) a patient with a mutated CALR.
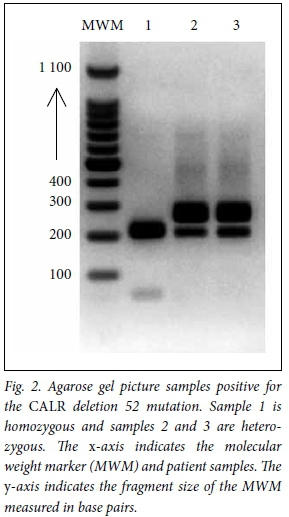
DNA fragments separated on a 2.5% agarose gel confirmed three samples positive for a 52-base pair deletion. Again, of the three samples, two were heterozygous and one was homozygous for a 52-base pair deletion in CALR. Fig. 2 shows a gel electropherogram of all samples positive for a CALR type 1 mutation. Included is a molecular weight marker sizing the fragments between 200 and 300 base pairs. The online program Lalign allowed for comparison of a reference sequence of CALR with the sequencing product. All three samples showed a 52-base pair deletion.
Discussion
CALR mutations were identified in three patients, all of which accounted for a type 1 mutation. Two were heterozygous for a 52-base pair deletion, while the other was homozygous. The JAK2 and CALR mutations were thought to be mutually exclusive, but the homozygous sample was double-positive for both JAK2 and CALR. Studies have described similar cases in patients with ET, but not in those with PV, and the implications relating to clinical presentation and prognosis of these patients are unclear. The heterozygous samples were from patients with a typical ET phenotype, and the homozygous sample had a PV phenotype.
The loss of CALR function can be observed by the onset of disease due to a frameshift to an alternative reading frame occurring in the C-terminus, obscuring CALR's pleotropic role. The C-terminus is coded by exon 9 of CALR, which includes an ER retention signal. With the generation of a new C-terminus, through replacement with a positively charged polypeptide chain rich in arginine and methionine, the ER KDEL retention signal is lost. CALR's roles are related to its distribution in the ER, in cytosol and on the cell surface. It resides mainly in the ER, where it performs its pleotropic functions ranging from Ca2+ regulation to ensuring proper protein folding. If the ER KDEL retention signal is lost, CALR therefore cannot be localised, or perform its critical functions in many biological processes including proliferation, apoptosis and immunogenic cell death.[10] When CALR is overexpressed, it suppresses cell proliferation and enhances apoptosis, which is triggered by Ca2+ release from the ER. Disabled cellular responses are therefore seen when there is a loss of CALR. In addition, it promotes the phagocytic function of macrophages to engulf hazardous cancerous cells. When CALR is mutated, there is therefore decreased phagocytosis of cancer cells.[3]
The unanticipated functional involvement of the JAK/STAT proteins signalling pathway alongside CALR demonstrates a transcriptional JAK2 signalling signature consistent in all MPN patients regardless of clinical phenotypes or mutational status. [3] This signature, present in patients with a mutated CALR, could elucidate the double-positive cases. Cells expressing type 1 CALR mutations proliferate independently of interleukin 3 (IL-3). This hypersensitivity to IL-3 hypothesis was confirmed by the phosphorylation of STAT5 in the presence and absence of IL-3. Increased phosphorylation of STAT5 was observed in the absence of IL-3, supporting the hypothesis that the JAK/STAT signalling pathway is activated in type 1 CALR mutations.[10]
Some data also suggest preferential expansion of the megakaryocyte cell lineage as a result of CALR's inability to export Ca2+ from the ER, keeping the calcineurin-nuclear factor of activated T cells (NFAT) signalling pathway notably less active. This in turn favours the megakaryocyte cell lineage instead of the erythroid lineage, providing an explanation as to why two heterozygous samples were observed in patients with high platelet counts. CALR expression in erythroblasts and granulocytes is also downregulated from CD34+ cells, suggesting CALR's crucial role in platelet functions.[5] In contrast to mutant CALR only being observed in ET and PMF, the homozygous sample was found in a patient suspected of having PV.
Conclusions
The clinical outcomes of CALR differ from those of JAK2 and MPL. CALR mutations are associated with prognostic advantages such as a lower thrombotic risk, longer median survival, and a more benign course of disease.[3] Since CALR mutations are associated with better outcomes, these clinical outcomes are useful in future prognostic and therapeutic indications of Ph chromosome-negative disorders. Knowledge of CALR mutations can therefore allow for a more definitive classification of myeloproliferative neoplasms.
Funding. This study was funded by the Department of Haematology and Cell Biology in the Faculty of Health Sciences at the University of the Free State.
References
1. Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009;114(5):937-951. http://dx.doi.org/10.1182/blood-2009-03-209262 [ Links ]
2. Klampfl T, Gisslinge H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;369(25):2379-2390. http://dx.doi.org/10.1056/NEJMoa1311347 [ Links ]
3. Luo W, Yu Z. Calreticulin (CALR) mutation in myeloproliferative neoplasms (MPNs). Stem Cell Invest 2015;2(16). http://dx.doi.org/10.3978/j.issn.2306-9759.2015.08.01 [ Links ]
4. Yokoyama M, Hirata KI. New function of calreticulin: Calreticulin-dependent mRNA destabilization. Circ Res 2005;97(10):961-963. http://dx.doi.org/10.1161/01.RES.0000193564.46466.2a [ Links ]
5. Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J 1999;344(2):281-292. http://dx.doi.org/10.1042/0264-6021:3440281 [ Links ]
6. Mendlovic F, Conconi M. Calreticulin: A multifaceted protein. Nature Educ 2010;4(1):1. [ Links ]
7. Tefferi A, Wassie EA, Guglielmelli P, et al. Type 1 versus type 2 calreticulin mutations in essential thrombocythemia: A collaborative study of 1027 patients. Am J Hematol 2014;89(8):E121-E124. http://dx.doi.org/10.1002/ajh.23743 [ Links ]
8. Ensembl. Gene: CALR ENSG00000179218. 2015. http://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000179218;r=19:12938578-12944489 (accessed 15 April 2015). [ Links ]
9. Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math 1991;12(3):337-357. http://dx.doi.org/10.1016/0196-8858(91)90017-D [ Links ]
10. Lavi N. Calreticulin mutations in myeloproliferative neoplasms. Rambam Maimonides Med J 2014;5(4):1-8. http://dx.doi.org/10.5041/RMMJ.10169 [ Links ]
 Correspondence:
Correspondence:
A de Kock
dekocka@ufs.ac.za
Accepted 16 March 2016.














