Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.106 n.10 Pretoria Oct. 2016
http://dx.doi.org/10.7196/samj.2016.v106i10.11300
IN PRACTICE
CLINICAL UPDATE
Molecular detection of carbapenemase-producing genes in referral Enterobacteriaceae in South Africa: A short report
O PerovicI, II; E BritzIII; V ChettyIV; A Singh-MoodleyV
IMD, DTM&H, FCPath (SA) (Micro), MMed (Micro), Centre for Opportunistic, Tropical and Hospital Infections, National Institute for Communicable Diseases, Johannesburg, South Africa
IIMD, DTM&H, FCPath (SA) (Micro), MMed (Micro), Department of Clinical Microbiology and Infectious Diseases, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
IIIMB ChB, MPH, Centre for Opportunistic, Tropical and Hospital Infections, National Institute for Communicable Diseases, Johannesburg, South Africa
IVMPH, Centre for Opportunistic, Tropical and Hospital Infections, National Institute for Communicable Diseases, Johannesburg, South Africa
VBSc, BMedSc Hons, MMedSc, PhD, Centre for Opportunistic, Tropical and Hospital Infections, National Institute for Communicable Diseases, Johannesburg, South Africa
ABSTRACT
Molecular confirmation of carbapenemase-producing Enterobacteriaceae (CPE) was introduced in South Africa (SA) at the end of 2011. We report on the detection of these resistance genes based on referral isolates. Enterobacteriaceae with non-susceptibility to any of the carbapenems according to defined criteria for antimicrobial susceptibility testing results were sent to a reference laboratory. A proportion of isolates had limited demographic, epidemiological and clinical data available. Organism identification was reconfirmed using reference laboratory methods, and the presence of carbapenemases was confirmed with a real-time polymerase chain reaction. We analysed 1 503 significant isolates received for confirmation from the National Health Laboratory Service and some private laboratories during 2012 - 2015 and confirmed one or more carbapenemase-producing genes in 68% of isolates, the most common organism being Klebsiella pneumoniae (60%). The most common carbapenemase genes were blaNDM, followed by blaOXA-48 and its variants. BlaOXA-48 and its variants demonstrated non-susceptibility to ertapenem in 89% of the isolates when analysed by the phenotypic method, and to ceftazidime in 34%. Overall, the detection rate for carbapenemases in K. pneumoniae blood isolates in the public sector was 1.9% during the 4-year period. This report indicates the presence of CPE in SA, and it is important for all healthcare workers to be aware of this major public health threat so that infection prevention and control measures can be implemented to prevent the spread of CPE in healthcare facilities.
Enterobacteriaceae are a large group of Gram-negative, rod-shaped bacteria. They are the bacteria most frequently isolated from clinical specimens and may account for up to 80% of all clinically significant isolates from Gram-negative bacilli and up to 50% of all clinically significant bacteria.[1] Enterobacteriaceae cause both nosocomial and community-acquired infections and are increasingly becoming multidrug resistant (MDR) to antimicrobial agents. The past few decades have seen the rapid emergence and spread of antimicrobial resistance, with clinicians having to rely on the carbapenem class of antibiotics to treat these resistant organisms. [2] However, increasing rates of carbapenem resistance are being reported.[3] The mechanism of resistance to carbapenems among Enterobacteriaceae is complex and mediated by several different mechanisms, such as the over-production of ampC enzymes, extended-spectrum beta-lactamases (ESBLs), carbapenemases that inactivate the ß-lactam antibiotics, including the carbapenems, efflux pumps and deletion of porins.[3-5] There are several classes of carbapenemase-produ-cing Enterobacteriaceae (CPE), which include non-metalloenzymes (Klebsiella pneumoniae carbapenemases (KPC), Guiana extended-spectrum ß-lactamases (GES), oxacillinase-type carbapenemases (OXA-48) and their derivatives) and metallo-ß-lactamases (MBL) (imipenemases (IMP), Verona integron-encoded MBL (VIM) and New Delhi metallo-ß-lactamase (NDM-1/2)). CPE are associated with increased morbidity and mortality as a result of limited treatment options rather than the expression of specific virulence characteristics.[6,7] CPE also have the potential for widespread transmission of carbapenem resistance owing to easily transmissible resistance genes on plasmids and chromosomes.[6,7] The focus given to carbapenemase detection despite all other mechanisms of resistance was due to the availability of molecular test methods.
Among all CPE, the most common enzyme was NDM-1.[3] NDM-1 was first described in 2008 in a Swedish patient returning from New Delhi, India,[8] in both Escherichia coli and K. pneumoniae isolates, which carried the novel MBL gene (blaNDM-1).[9]NDM-1 has subsequently been reported worldwide, with most early cases of NDM-1 diagnosed in the UK having epidemiological links with the Indian subcontinent.[8]
Objective
To demonstrate the presence of carbapenemases in Enterobacteri-aceae over a 4-year period, based on a referral system for confirmation of CPE genes.
Methods
Carbapenem non-susceptible, clinically significant isolates from the Enterobacteriaceae family were submitted to the Antimicrobial Resistance Laboratory (AMRL) at the National Institute for Communicable Diseases (NICD), Johannesburg, South Africa (SA), for confirmation of carbapenemase-producing genes from 2012 through 2015. Referral of isolates by public and private microbiology laboratories was based on non-susceptibility to carbapenems by disc diffusion (Kirby-Bauer method, using Oxoid Antimicrobial Susceptibility Test Discs (Oxoid, UK)) or minimal inhibitory concentration (MIC) testing methods interpreted by the Clinical and Laboratory Standards Institute guideline.[10] Submission practice from public laboratories was based on a guideline from the reference laboratory at the NICD and from private laboratories on a voluntary basis.
Organism identification was reconfirmed using automated systems (VITEK 2 (bio-Mêrieux, France) and/or Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-ToF, Bruker Daltonik GmbH, Germany), and antimicrobial susceptibility testing (AST) was done using the MicroScan Walkaway system (Siemens, USA) at the AMRL. For molecular methods, DNA was extracted from purity plates using a crude boiling method at 95oC for 25 minutes. The supernatant was harvested and screened for blaNDM, blaKPC, blaOXA-48 and its variants, blaGES, blaIMP and blaVIM, using a real-time polymerase chain reaction (LightCycler 480 II, Roche Applied Science, Germany), the LightCycler 480 Probes Master kit (Roche Diagnostics, USA) and individual LightMix Modular kits (Roche Diagnostics, USA). Kit-positive controls as well as in-house controls were used in all assays (blaNDM, ATCC BAA21246; blaKPC, ATCC BAA1705; blaOXA-48 and its variants, NCTC13442; blaGES, clinical isolate; blaIMP, NCTC 13476; and blaVIM, clinical isolate). Sterile water was used as a negative control.
Statistical analysis
Where appropriate, we calculated frequencies and percentages and used the χ2 test/Fisher's exact test to compare categorical variables. Statistical analyses were performed using STATA 14 (StataCorp, USA).
Ethical considerations
Laboratory-based antimicrobial resistance surveillance for nosocomial bacteria was approved by the Human Research Ethics Committee (Medical) at the University of the Witwatersrand, Johannesburg (clearance certificate no. M10464).
Results
We analysed 1 503 clinically significant Enterobacteriaceae isolates received for carbapenemase-producing gene confirmation. The age distribution of patients with carbapenem-resistant Enterobacteriaceae (CRE) infections showed two significant peaks, in children aged 0 - 5 years and in adults aged 30 - 40 years (p=0.05). Blood was the most common specimen type (25%), followed by urine (22%). We confirmed ESBL in 93% of isolates by the automated MIC method. Furthermore, carbapenemase-producing genes were confirmed molecularly in 68% of isolates (1 021/1 503). Of all Enterobacteriaceae, the most common was K. pneumoniae (60%), followed by Enterobacter cloacae (14%) and Serratia marcescens (6%).
The most common carbapenemase-pro-ducing genes were blaNDM, followed by blaOXA-48 (Fig. 1). Carbapenemases were confirmed in 672 K. pneumoniae isolates (68%), and blaNDMwas the most common gene identified (58%).
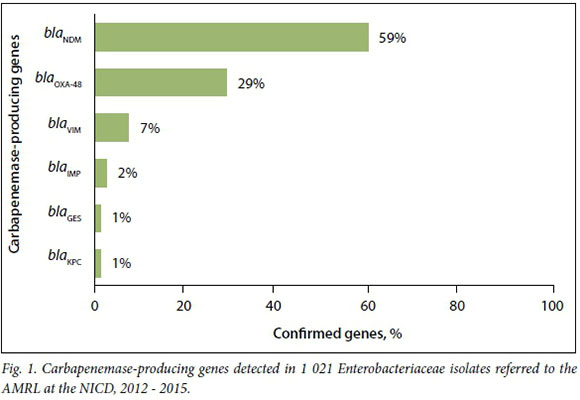
Of the blaNDM-positive isolates, 0.8% and 0.6% showed susceptibility to ertapenem and ceftazidime, respectively. BlaOXA-48 and its derivatives demonstrated susceptibility to ertapenem in 11% of isolates and to ceftazidime in 66%.
The detection rate of carbapenemases among all K. pneumoniae blood isolates from public laboratories during the 4-year period was 1.9%. Based on voluntary referral practice, we could not estimate the prevalence of carbapenemases for all Enterobacteriaceae isolates and specimen types in SA.
Discussion
This short report describes emerging resistance to carbapenems in Enterobacteri-acae over a 4-year period in SA. We detected the presence of major carbapenemases, i.e. NDM, OXA-48 and VIM. In a previous laboratory-based antimicrobial resistance surveillance study conducted in 2010 - 2012, no isolates containing blaNDM or blaKPC were found, and blaGESand blaVIMwere confirmed in <0.1%.[11] The present report suggests rapid dissemination of these genes once they are introduced into the environment, and we describe its longitudinal nature, which is in line with the global dissemination.[12]
The vast majority of the referral isolates produced ESBL, which is to be expected owing to the MDR patterns of these organisms and compares with a previous surveillance report.[11] Enterobacteriaceae extensively exhibit MDR patterns, which enable them to persist and spread rapidly in healthcare settings.[13]
Our results indicate that blaOXA-48-positive isolates could be missed in 11% of isolates owing to susceptibility to ertapenem, yet 43% were sensitive to imipenem and 57% to meropenem, which demonstrated that AST methods cannot be used for screening of these enzymes. This differs from an Indian study that showed 75% sensitivity to ertapenem and 84% sensitivity to imipenem and meropenem.'141 Poirel et al.'151pointed out menace behaviour of these enzymes and the difficulties this poses for phenotypic detection. As we report here, some of these enzymes will be overlooked, particularly if carbapenems are used as indicators for AST resistance screening.
There are various approaches to the control of MDR organisms. Carmeli et al.[16]indicated that the objective for control should be eradication, while others such as Thurlow et al.[17]considered that a wider approach is needed once endemicity is established. Thurlow et al.[17]also suggested that reducing the burden of CPE on patients' skin should be explored further as a way of reducing cross-transmission at long-term healthcare hospitals, where endemicity is most likely. Clinicians and infection control practitioners should be aware of the presence of carbapenemases in Enterobacteriaceae and its implications for infection prevention and control in the SA setting.
Study limitations
This report has a number of limitations. Owing to lack of policies and voluntary practice in sending isolates for confirmation to the reference laboratory, we were unable to determine the national prevalence of CPE or to establish whether there has been an increase in CPE in SA. Changes in submission practices and/or increased awareness of CPE infections undoubtedly influenced the number of isolates referred to the reference laboratory. However, this analysis reports a minimum estimate of the presence of CPE organisms and carbapenemase-producing genes in the country. Missing demographic, epidemiological and clinical data reduced our ability to analyse laboratory data in more meaningful ways. Surveillance for CPE through the Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) surveillance platform was introduced at 12 sentinel sites in four SA provinces in 2015. Future analysis of surveillance data for CPE should provide a more representative estimate of their prevalence and distribution in SA.
Conclusions
This report indicates the presence of CPE in SA. It is essential for all clinicians to be aware of this major public threat and be prepared to act if CPE occurs in patients. At all healthcare facilities, the importance of enforcing infection prevention and control measures to prevent the spread of CPE should be emphasised. Importantly, antimicrobial stewardship programmes should be implemented at facility level to prevent selection pressure on bacterial organisms to develop resistance.
Acknowledgements. We thank all members of the AMRL at the NICD, and particularly Rubeina Badat, Naseema Bulbulia and Ruth Mohlabeng for their technical support. We also thank Penny Crowther for database design.
REFERENCES
1. Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. Enterobacteriaceae: Introduction and identification. In: Farmer JJ III, ed. Manual of Clinical Microbiology. Philadelphia: Elsevier, 2003:647. [ Links ]
2. Jacob JT, Klein E, Laxminarayan R, et al. Vital signs: Carbapenem-resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep 2013;62(9):165-170. [ Links ]
3. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011;17(10):1791-1798. DOI:10.3201/eid1710.110655 [ Links ]
4. Spellberg B, Blaser M, Guidos RJ, et al. Combating antimicrobial resistance: Policy recommendations to save lives. Clin Infect Dis 2011;52(Suppl 5):S397-S428. DOI:10.1093/cid/cir153 [ Links ]
5. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2012;18(9):1503-1507. DOI:10.3201/eid1809.120355 [ Links ]
6. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 2008;29(12):1099-1106. DOI:10.1086/592412 [ Links ]
7. Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001;45(4):1151-1161. DOI:10.1128/AAC.45.4.1151-1161.2001 [ Links ]
8. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009;53(12):5046-5054. DOI:10.1128/AAC.00774-09 [ Links ]
9. Brink AJ, Coetzee J, Clay CG, et al. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J Clin Microbiol 2012;50(2):525-527. DOI:10.1128/JCM.02234-12 [ Links ]
10. Patel JB, Cockerill FR, Eliopoulos GM, et al. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Wayne, Penn., USA: Clinical and Laboratory Standards Institute, 2016. [ Links ]
11. Perovic O, Singh-Moodley A, Duse AG, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010 - 2012. S Afr Med J 2014;104(8):563-568. DOI:10.7196/SAMJ.7617 [ Links ]
12. Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: Clinical perspectives on detection, treatment and infection control. J Intern Med 2015;277(5):501-512. DOI:10.1111/joim.12342 [ Links ]
13. Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin Microbiol Rev 2012;25(4):682-707. DOI:10.1128/CMR.05035-11 [ Links ]
14. Sugumar M, Kumar KM, Manoharan A, Anbarasu A, Ramaiah S. Detection of OXA-1 beta-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS One 2014;9(3):e91800. DOI:10.1371/journal.pone.0091800 [ Links ]
15. Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum beta- lactamases in Gram-negative rods. Infect Genet Evol 2012;12(5):883-893. DOI: 10.1016/jmeegd.2012.02.008 [ Links ]
16. Carmeli Y, Akova M, Cornaglia G, et al. Controlling the spread of carbapenemase-producing Gram- negatives: Therapeutic approach and infection control. Clin Microbiol Infect 2010;16(2):102-111. DOI:10.1111/j.1469-0691.2009.03115.x [ Links ]
17. Thurlow CJ, Prabaker K, Lin MY, Lolans K, Weinstein RA, Hayden MK. Anatomic sites of patient colonization and environmental contamination with Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae at long-term acute care hospitals. Infect Control Hosp Epidemiol 2013;34(1):56-61. DOI:10.1086/668783 [ Links ]
 Correspondence:
Correspondence:
O Perovic
olgap@nicd.ac.za
Accepted 26 July 2016.














