Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.106 n.1 Pretoria Jan. 2016
http://dx.doi.org/10.7196/SAMJ.2016.V106I1.9571
RESEARCH
HLA typing: Conventional techniques v. next-generation sequencing
J MelletI; C M GrayII, III; M S PepperIV
IBSc Hons, MSc Department of Immunology, Institute for Cellular and Molecular Medicine, and South African Medical Research Council Extramural Unit for Stem Cell Research and Therapy, School of Medicine, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa
IIBSc Hons, MSc, PhD; Division of Immunology, Institute of Infectious Diseases and Molecular Medicine and Clinical Laboratory Sciences, Faculty of Health Sciences, University of Cape Town, South Africa
IIIBSc Hons, MSc, PhD; Laboratory for Tissue Immunology, National Health Laboratory Services, Groote Schuur Hospital, Cape Town, South Africa
IVMB ChB, PhD, MD, Department of Immunology, Institute for Cellular and Molecular Medicine, and South African Medical Research Council Extramural Unit for Stem Cell Research and Therapy, School of Medicine, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa
ABSTRACT
BACKGROUND. The large number of population-specific polymorphisms present in the HLA complex in the South African (SA) population reduces the probability of finding an adequate HLA-matched donor for individuals in need of an unrelated haematopoietic stem cell transplantation (HSCT). Next-generation sequencing (NGS) has numerous advantages compared with conventional typing techniques.
OBJECTIVE. To evaluate whether NGS can provide any additional value over conventional techniques in the SA context for the purpose of HSCT and cord blood banking.
METHODS. HLA genotyping was performed using NGS on 20 samples that had previously been HLA typed by conventional methods to evaluate whether NGS might provide any additional value over conventional HLA determination techniques.
RESULTS. NGS of routinely sequenced loci and exons yielded accurate genotypes for 98.5% of the five loci of interest, compared with 98% when additional exons were included.
CONCLUSION. The study shows that the additional value of NGS over conventional techniques is limited, and unless done on a large scale to reduce cost may not be appropriate in SA at this stage in the context of HSCT and cord blood banking.
The HLA complex, located on chromosome 6, comprises the most polymorphic genes in humans[1] and plays a pivotal role in matching for haematopoietic stem cel1 transplantation (HSCT).[2] Allele-level HLA matching between donors and recipients reduces the likelihood of rejection and graft-versus-host disease (GVHD).[3] The South African (SA) population is characterised by great genetic diversity and the presence of population-specific and uncommon alleles decreases the probability of finding an HLA-compatible donor. The majority of individuals in a given population group possess common alleles, but several uncommon population-specific alleles are also present.[4]
HLA typing was initially performed using serological techniques. In the 1960s, this was the sole method of determining tissue types. Even though this method is still performed in some laboratories today, there are numerous limitations. In the mid-1990s, DNA-based techniques became more popular and were used to complement serological techniques. Today most laboratories primarily use probe/ primer-based techniques, which assign genotypes on the basis of previously identified alleles. However, as a result of the ever- increasing number of new alleles, genotyping has become challenging. An accurate high-resolution HLA genotyping method is therefore a necessary tool for the matching of donors to patients in need of an unrelated HSCT. Inaccurate typing could lead to inadequate HLA matching between donors and recipients, which could ultimately increase the chances of graft rejection, GVHD and mortality.
The existing techniques have contributed significantly to our current knowledge of allelic diversity. At present, sequence-based typing (SBT) methods, in particular next-generation sequencing (NGS), provide the highest possible resolution. NGS platforms were initially only used for genomic sequencing, but also showed potential for research and diagnostic purposes. Even though these newly developed techniques have already proved to be efficient in identifying novel alleles, the more conventional techniques are still preferred for routine procedures in many diagnostic laboratories. Exons 2 and 3 for class I and exon 2 for class II are routinely sequenced because they constitute the peptide-binding region of the corresponding HLA molecules. Alleles that are identical across this region but differ in other exons are referred to as ambiguous alleles. Sequencing of additional exons has been shown to reduce these ambiguities and produce better allele resolution, and could improve matching between unrelated donors and recipients for HSCTs.[5]
The purpose of this study was to evaluate whether NGS can provide any additional value over conventional techniques in the SA context for the purpose of HSCT and cord blood banking.
Methods
Twenty DNA samples isolated from peripheral blood mononuclear cells were selected from the South African Bone Marrow Registry. Each DNA sample had already been HLA genotyped by the Laboratory for Tissue Immunology at the time of commencement of the study. HLA typing was performed using low- and/or high- resolution typing techniques. This study made use of the Life Sciences, Roche 454 NGS platform for genotyping of the samples. All laboratory procedures for this study were performed according to the HLA assay manual (Roche Applied Science GS GHLA Assay Manual, March 2011). The typing kit targets the most hypervariable regions of the MHC class I and class II genes. The GS GType HLA Primer sets (Roche Applied Science, Germany) were made available as two kits, medium resolution (MR) and high resolution (HR). Sequencing was performed by Inqaba Biotec on a GS Junior sequencer. The raw sequencing data were assembled and analysed using JSI SeqHLA 454 software (version 3.16.0) (JSI Medical Systems, Germany).
Ethical considerations
This study was conducted in the Department of Immunology at the University of Pretoria, SA. Ethical approval was granted in 2010 by the Faculty of Health Sciences Research Ethics Committee (Protocol No. 131/2010) for the project entitled 'Feasibility study for a public cord blood stem cell bank in South Africa of which the current study formed part. The proposal for this study was submitted in November 2011, followed by ethical approval, which was granted by the Ethics Committee of the University of Pretoria (Protocol No. 219/2011). Separate ethical approval was granted by the University of Cape Town (Protocol No. 523/2011) for the use of the 20 samples obtained from the Laboratory for Tissue Immunology, Groote Schuur Hospital, Cape Town.
Results
Samples 1 - 10 (Table 1) were previously typed by high resolution at class I and class II loci. Samples 11 - 20 (Table 2) were typed by low resolution at class I loci and by high resolution at class II loci. Genotypes obtained using these conventional techniques are shown in column 3, and the results obtained from the present study by NGS (Roche 454) of routinely sequenced exons and additional exons for the five loci of interest are shown in columns 4 and 5, respectively. Results from the conventional techniques are displayed at a two- to four-digit level of resolution, depending on the technique used. In cases where a conventional typing result was not identical to an allele in the ambiguity list obtained by NGS, a sample was said to be in disagreement.
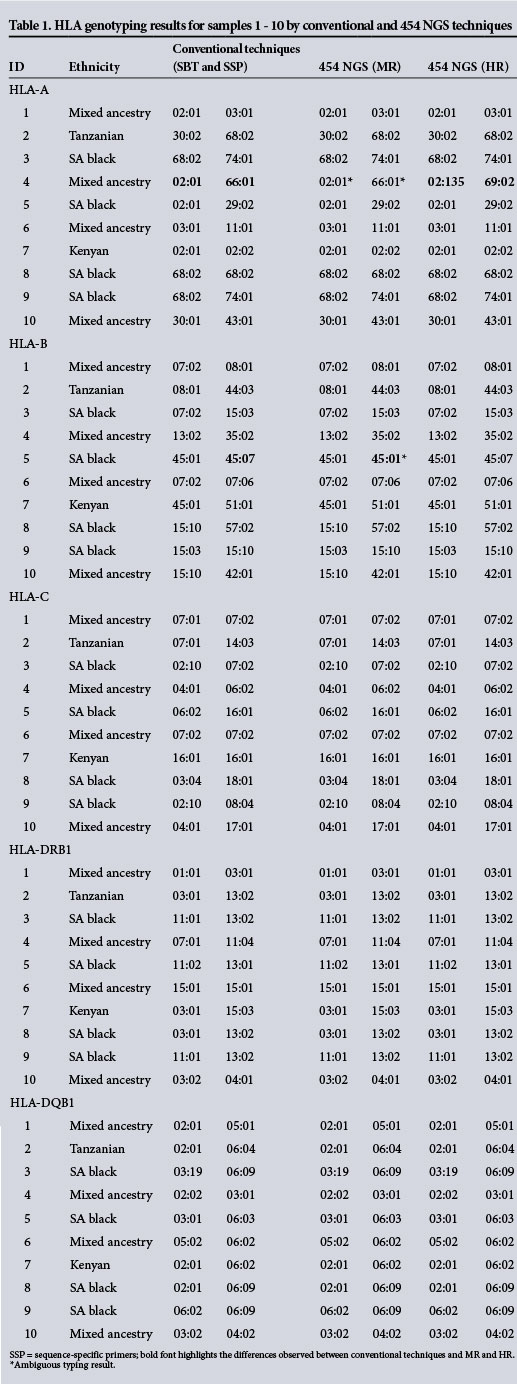
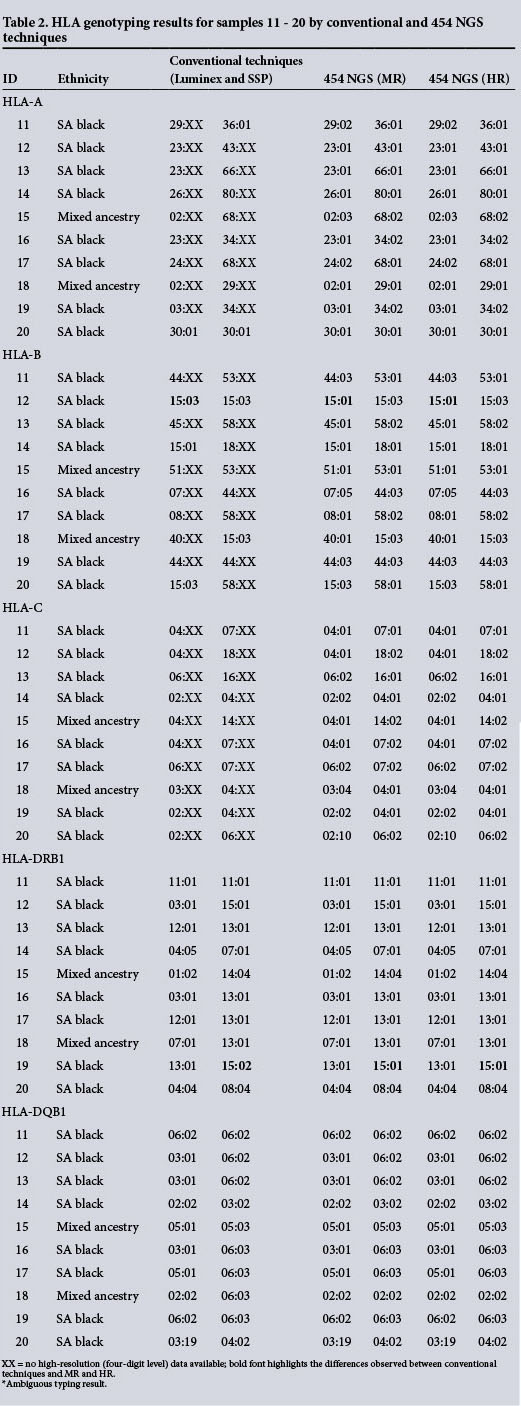
Side-by-side comparison of the results obtained from conventional and NGS typing for samples 1 - 10 showed 99% and 98% concordance between conventional typing techniques and NGS for routinely sequenced exons and additional exons, respectively. The results for samples 11 - 20 showed 98% concordance between conventional typing techniques and NGS for both routinely sequenced exons and additional exons. It was possible to assign accurate genotypes to 98.25% of the loci of interest for the 20 samples by NGS.
The genotypic discordance between the conventional techniques and NGS typing of routinely sequenced exons was mainly due to ambiguous typing results. Many alleles are identical across exons 2 and 3 of the HLA genes, since several polymorphisms are located outside the sequenced region. [6] Discordance was observed for samples 4 (HLA-A in an individual of mixed ancestry), 5 (HLA-B in a black South African), 12 (HLA-B in a black South African) and 19 (HLA-DRB1 in a black South African).
Discussion
HLA genotyping is performed on a routine basis for various applications including HSCT. Mismatching between donors and recipients could lead to graft rejection and increased morbidity and mortality. Accurate HLA typing is therefore critical for successful engraftment. The limited number of studies that have targeted African populations and the high diversity of these individuals affect the degree of certainty with which a genotype is assigned. In many instances, genotypes are assigned based on the predominant frequencies of HLA genotypes in a given population. This could affect the assignment of rare alleles, especially in African populations, where many alleles have not yet been comprehensively described. Numerous alleles are identical across exons 2 and 3 of the HLA genes,[6] which creates a challenge in accurately assigning HLA genotypes, leading to ambiguous typing results. The ambiguity observed in this cohort is greater when compared with the results of Holcomb et al.[7] The degree of ambiguity observed could be a result of the paucity of knowledge on polymorphisms at HLA loci present in black SA and African individuals in general. An alternative approach to resolving ambiguity would be to sequence the entire gene of interest,[8] which might be helpful in resolving this ambiguity. However, polymorphisms outside the peptide-binding region may not affect the outcome of transplantation. According to a study by Pasi et al.,[9]several DRB1 alleles are identical across the peptide-binding region, but possess nucleotide changes outside the peptide-binding region that are unlikely to influence the outcome of transplantation.
The degree of resolution obtained for HLA typing has increased over the years with the emergence of various DNA-based typing techniques. Serological techniques are able to assess antigen expression but are unable to distinguish between cross- reactive groups. Probe- and primer- based DNA typing methods are able to determine alleles based on known variants. However, rare and undescribed variants cannot be identified by these techniques, which creates a challenge in accurately
assigning unknown HLA genotypes. The primer-based method has higher specificity and is able to genotype at an intermediate resolution. As more HLA alleles are discovered, both these techniques require updated primers and probes to account for the allelic diversity present in a given population. The degree of HLA variation found in African populations makes it challenging to assign genotypes typed at low to intermediate resolution. It is therefore essential that a reliable minimal four-digit resolution typing method be used for correct assignment of HLA alleles and haplotypes, especially for HSCT. The question is whether SBT or NGS technologies enable better resolution of HLA ambiguities, especially in African populations. The data in this small sample set suggest that neither method has the advantage over the other, and that 454 NGS, despite generating large numbers of shorter reads, does not provide a great enough increment in resolution to warrant implementation on a routine basis.
Conclusion
The equipment and reagents for NGS techniques are costly and not readily accessible to the majority of research and diagnostic institutions in developing countries such as SA. This study therefore indicates that the value of NGS over conventional techniques will only become significant in the context of HSCT and cord blood banking in SA when the number of samples increases to the point where NGS becomes more cost-effective than conventional techniques.[10]
Acknowledgements. This research was funded by the South African Medical Research Council in terms of the MRC's Flagship Award Project SAMRC-RFA-UFSP-01-2013/STEM CELLS, the National Research Foundation Internship Programme and the Institute for Cellular and Molecular Medicine of the University of Pretoria. We are grateful to Roche Diagnostics (SA and USA) for the technical support in the laboratory and data analysis.
References
1. A haplotype map of the human genome. Nature 2005;437(7063):1299-1320. [http://di.doi.org/10.1038/nature04226] [ Links ]
2. Fürst D, Müller C, Vucinic V, et al. High-resolution HLA matching in hematopoietic stem cell transplantation: A retrospective collaborative analysis. Blood 2013;122(18):3220- 3229. [http://di.doi.org/10.1182/blood-2013-02-482547] [ Links ]
3. Kanda Y, Kanda J, Atsuta Y, et al. Changes in the clinical impact of high-risk human leukocyte antigen allele mismatch combinations on the outcome of unrelated bone marrow transplantation. Biol Blood Marrow Transplant 2014;20(4):526- 535. [http://dx.doi.org/10.1016/j.bbmt.2014.01.003] [ Links ]
4. Paximadis M, Mathebula TY, Gentle NL, et al. Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Hum Immunol 2012;73(1):80- 92. [http://di.doi.org/10.1016/j.humimm.2011.10.013] [ Links ]
5. Ehrenberg PK, Geretz A, Baldwin KM, et al. High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics 2014;15(1):864. [http://dx.doi.org/10.1186/1471-2164-15-864] [ Links ]
6. Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res 2015;43(Database Issue):D423-D431. [http://dx.doi.org/10.1093/nar/gku1161] [ Links ]
7. Holcomb CL, Höglund B, Anderson MW, et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens 2011;77(3):206-217. [http://dx.doi.org/10.1111/j.1399-0039.2010.01606.x] [ Links ]
8. Erlich RL, Jia X, Anderson S, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics 2011;12(1):42. [http://dx.doi.org/10.1186/1471-2164-12-42] [ Links ]
9. Pasi A, Crocchiolo R, Bontempelli M, et al. The conundrum of HLA-DRB1»14:01/»14:54 and HLA-DRB3»02:01/»02:02 mis- matches in unrelated hematopoietic SCT. Bone Marrow Transpl 2011;46(7):916-922. [http://dx.doi.org/10.1038/bmt.2010.246] [ Links ]
10. Gabriel C, Fürst D, Faé I, et al. HLA typing by next-generation sequencing - getting closer to reality. Tissue Antigens 2014;83(2):65- 75. [http://dx.doi.org/10.1111/tan.12298] [ Links ]
 Correspondence:
Correspondence:
M S Pepper
michael.pepper@up.ac.za
Accepted 13 October 2015.














