Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.105 n.5 Pretoria May. 2015
http://dx.doi.org/10.7196/SAMJ.8183
RESEARCH
Community- versus healthcare-acquired bloodstream infections at Groote Schuur Hospital, Cape Town, South Africa
R McKayI; C BamfordII, III
IMSc; School of Population and Public Health, University of British Columbia, Vancouver, Canada
IIMB ChB, FCPath, MMed; Division of Medical Microbiology, University of Cape Town, South Africa
IIIMB ChB, FCPath, MMed; National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa
ABSTRACT
BACKGROUND: Bloodstream infections (BSIs) cause considerable morbidity and mortality. The epidemiology of bacterial infections differs in community and hospital settings. Regular surveillance and reporting of pathogens and antimicrobial susceptibility can assist in appropriate management of BSIs
OBJECTIVES: To describe the distribution of organisms and of antibiotic susceptibility among isolates from blood cultures at a tertiary academic hospital during a 1-year period, stratifying by place of infection acquisition
METHODS: This was a retrospective descriptive study of bloodstream isolates from cultures from adults (>13 years of age) routinely submitted between 1 October 2011 and 30 September 2012 to the clinical laboratory at Groote Schuur Hospital, Cape Town, South Africa. Community-acquired infections were compared with healthcare-acquired infections, defined as infections developing at least 48 hours after admission or within 3 months of admission to a healthcare facility. Frequencies and proportions of infecting organisms are presented, along with susceptibility results for selected pathogens. The hospital-acquired isolates were stratified by ward (emergency, general medical or general surgical ward or intensive care unit (ICU)) to determine organism frequency and susceptibility patterns by hospital ward
RESULTS: Among adults, 740 non-duplicate pathogens were isolated from BSIs. Nearly three-quarters of infections were healthcare acquired. Enterobacteriaceae and non-fermentative Gram-negative bacilli were predominant among healthcare-acquired pathogens (39.2% and 28.5%, respectively), while Enterobacteriaceae and Gram-positive organisms were the most common among community-acquired pathogens (39.2% and 54.3%, respectively). The majority of community-acquired Enterobacteriaceae were highly susceptible to antibiotics (gentamicin 95.6%, ceftriaxone 96.1% and ciprofloxacin 92.2%), whereas 64.6% of healthcare-associated isolates were susceptible to gentamicin, 58.5% to ceftriaxone and 70% to ciprofloxacin. All community-acquired Staphylococcus aureus isolates v. 52.4% of healthcare-acquired isolates were susceptible to cloxacillin. The susceptibility of healthcare-acquired Pseudomonas aeruginosa and Acinetobacter baumanii complex isolates was <80% to all antibiotics with the exception of colistin. Klebsiella spp., S. aureus and Escherichia coli were the commonest causes of healthcare-acquired infections in all areas outside of the ICUs, whereas Acinetobacter was common in the ICUs and rare in all other areas
CONCLUSION: The distinction between community- and healthcare-acquired infections is critical in antibiotic selection because narrow-spectrum agents can be utilised for community-acquired infections. The considerable antibiotic resistance of healthcare-acquired pathogens highlights the importance of infection prevention and control. This type of surveillance could be incorporated into routine laboratory practice
Bloodstream infections (BSIs) cause considerable morbidity and mortality.[1,2] Estimates suggest that 10 - 13% of community-onset BSIs are fatal,[3,4] while 23% of nosocomial BSIs resulted in death in one study in the USA.[4] A paediatric cohort study in Kenya reported a case fatality rate of 24% for community-acquired and 53% for hospital-acquired bacteraemia.[5] A systematic review of admissions to hospital in various regions of Africa estimated that 13.5% of adults and 8.2% of children had community-acquired BSIs,[6] indicating these are a common cause of illness and account for a substantial proportion of all healthcare admissions. Rapid diagnosis, identification of the causative bacteria and appropriate treatment are essential in mitigating the morbidity and mortality associated with BSIs.
The epidemiology of bacterial infections differs in community and hospital settings. The predominant bacteria causing community-acquired infections are Gram-positive organisms, while hospital-acquired infections are more frequently caused by Gram-negative bacteria.[4] This distinction has relevance to empirical treatment of suspected bacterial infection.
In the past decade, an additional category of healthcare-associated infection has been recognised, to cover infections in patients who have had recent contact with the healthcare system.[7] Healthcare-associated infections are typically similar to hospital-acquired infections in terms of pathogens and susceptibility patterns. We have therefore chosen to combine hospital-acquired and healthcare-associated infections as healthcare-acquired infections.
Increasing antibiotic resistance complicates treatment of infections, in some cases seriously diminishing the options for effective therapy,[8] and is often associated with worse outcomes.[9] Laboratory-based surveillance data for BSIs in South Africa (SA) are available; however, these data do not distinguish between community- and hospital-acquired infection.[10] Regular surveillance and reporting of BSIs and antibiotic susceptibility, including differentiation of community- and healthcare-acquired infections, can assist in managing infections appropriately and in adapting local antibiotic stewardship policies.[11,12]
Groote Schuur Hospital (GSH) is a large tertiary academic and teaching hospital in Cape Town, SA. In this hospital, blood cultures are generally collected when a patient has clinical features of sepsis, typically before commencing or changing antibiotic therapy. However, there are no explicit guidelines or policies around indications for taking of blood cultures, and practices may vary from clinician to clinician.
This report describes the distribution of organisms isolated in blood cultures at GSH during a 1-year period. Additionally, we describe the distribution of antibiotic resistance among some groups of pathogens. We adapted routine laboratory practices to allow for classification of infections as healthcare or community acquired and applied this stratification to analysis of pathogens and antibiotic susceptibility results.
Guidelines to clinicians on antimicrobial therapy of bloodstream and other infections are provided by means of an annually updated and freely available booklet. At the time of the study, prior authorisation of most second-line antibiotics was required.
Methods
Study design
This was a retrospective assessment of bacterial isolates collected at GSH over a 12-month period.
Data
All microbiology testing was conducted at the GSH National Health Laboratory Service microbiology laboratory, which uses the BACTEC 9240 automated blood culture system (Becton Dickinson, USA). Identification and susceptibility testing was carried out using various standard methods employed in the laboratory. In the majority of cases standard biochemical tests and disc diffusion and gradient diffusion susceptibility tests were used for Gram-positive organisms, whereas a method of direct inoculation of the ID-GNB and the AST-N064 cards of the Vitek 2 system was used for Gram-negative organisms.[13] However, alternative methods were used if needed.
Susceptibility results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) criteria for the relevant year.[14,15] While the appropriate breakpoints for cephalosporins were utilised, the laboratory at this time, in line with the contemporary national practice, continued to accept the suggestion of the Vitek Advanced Expert System to report extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae as resistant to all cephalosporins. In addition, ESBL production was not reported on the laboratory information system (LIS) owing to limitations in the design and setup of the interface linking the Vitek instrument and the LIS. Hence, for the purposes of this analysis, ESBL production was assumed if Enterobacteriaceae isolates were reported as resistant to cefepime. Staphylococcus aureus isolates with an inducible clindamycin resistance phenotype were reported as resistant to clindamycin.
Culture result data were extracted for every bloodstream isolate from GSH recorded in the laboratory database for the period 1 October 2011 - 30 September 2012. To limit the data to single infection episodes, duplicates (defined as instances of an additional culture submitted from the same patient, isolating the same organism, within 14 days of a previous culture) were excluded.[5,16]
As part of regular practice, local microbiologists prospectively categorised each case of BSI after collecting relevant information from the attending clinician; this information was then recorded in a separate database, linkable by unique laboratory number.
Healthcare-acquired infections were defined as those in patients admitted to (any) hospital for at least 48 hours before developing signs and symptoms of infection, or who had been admitted to a healthcare facility within the 3 months preceding the date of blood culture. In addition, infections in patients living in a long-term care facility or receiving dialysis, chemotherapy or home-based intravenous therapy were also included as healthcare-acquired infections.[7] Community-acquired isolates were defined as those occurring before admission, or within the first 48 hours of admission in patients without recent hospitalisation in the past 3 months. Classification of isolates as contaminants was based on recognition of the organism as one commonly considered to be a contaminant (i.e. on the identity of the organism), provided this was supported by the treating clinician's clinical assessment and decision not to provide therapy targeted to the isolate.[17] In the majority of cases in our hospital only a single blood culture is submitted per episode of sepsis. However, if multiple cultures were submitted, the number of positive blood culture bottles or sets was also used to determine whether an isolate was categorised as a contaminant or not. Isolates for which no clinical information was available, usually owing to early discharge of the patient, were also classified as contaminants. Additional study variables included the date on which the blood sample was taken, the age and gender of the patient, and the hospital ward where the patient was located.
Ethical approval for the study was granted through the University of Cape Town's Faculty of Health Sciences Human Research Ethics Committee and the University of British Columbia's Clinical Research Ethics Board.
Analysis
We used descriptive methods to assess the distribution of bloodstream organisms and resistance patterns. Antibiotic susceptibility results were recorded in this database as susceptible, intermediate or resistant. Results are presented as the proportion of susceptible isolates out of the number of valid test results for that antibiotic and organism (or group of organisms). We further stratified the organism and susceptibility results by wards, grouped into categories of emergency, general medical, intensive care units (ICUs), maternity and surgery.
Fisher's exact tests were used to compare the proportions, with p<0.05 considered statistically significant. Data preparation and analysis were conducted in SAS software for Windows, version 9.3(SAS Institute, USA).
Results
Study sample
There were 1 730 blood culture records from adults >13 years of age in the database; of these, 799 were considered pathogens and 931 contaminants. One hundred and forty-three pathogens were removed as duplicates. After including multiple organisms isolated from individual blood cultures (87 additional pathogens), our dataset contained 740 pathogens, isolated from 653 episodes of BSI in 533 patients. There were 981 organisms isolated from 931 blood cultures that were considered contaminants.
Demographics
Seventy-three per cent of the 653 episodes of illness (n=472) and 73% of the 740 organisms isolated (n=543) were healthcare acquired. The mean patient age was 49.6 years (standard deviation (SD) 19.8) for those with community-acquired infections, and 45.5 years (SD 16.8) for those with healthcare-acquired infections (Table 1).

Microbiology
Among community-acquired isolates, Gram-positive organisms were most common (54.3%, n=107), while there were 77 Enterobacteriaceae isolates (39.1%) (Table 2). Non-fermenting Gram-negative bacilli and fungi were rare. Streptococcus pneumoniae was the most frequently isolated organism (21.3%, n=42), followed by Escherichia coli (19.8%, n=39) and S. aureus (15.2%, n=30).

Gram-negative bacilli constituted the largest group of healthcare-acquired isolates (68.0%, n=369) (Table 2). Gram-positive organisms and fungi accounted for 27.8% and 4.4%, respectively. Among the Gram-negative bacilli, Enterobacteriaceae predominated (39.4%, n=214), while non-fermenting Gram-negative bacilli constituted 28.4% (n=154). The most commonly isolated healthcare-acquired organisms were Acinetobacter baumanii complex (16.0%, n=87), Klebsiella spp. (15.8%, n=86), S. aureus (11.6%, n=63) and E. coli (9.9%, n=54).
Antibiotic susceptibility
Community-acquired isolates
Community-acquired bloodstream pathogens demonstrated minimal resistance to most first-line antibiotic agents used for treatment. All S. aureus isolates (n=30) were susceptible to cloxacillin, and 28 and 29 were susceptible to erythromycin and clindamycin, respectively (Fig. 1). The majority of S. pneumoniae isolates (35/42, 83%) were fully susceptible to penicillin (minimum inhibitory concentration (MIC) <0.06 μg/mL) while the remainder (7/42, 17%) had MICs in the range of 0.12 - 2 μg/ mL, meaning that they would be categorised as resistant in cases of meningitis, but susceptible for infections outside the central nervous system. All S. pneumoniae isolates were susceptible to ceftriaxone according to meningitis breakpoints (MIC <0.5 μg/mL).

The majority of Enterobacteriaceae were susceptible to gentamicin (95.6%), ceftriaxone (96.1%) and ciprofloxacin (92.2%) (Fig. 2). The proportion of ESBL-producing Enterobacteriaceae was just under 5%, and 79.1% were susceptible to amoxicillin/clavulanate.

Healthcare-acquired isolates
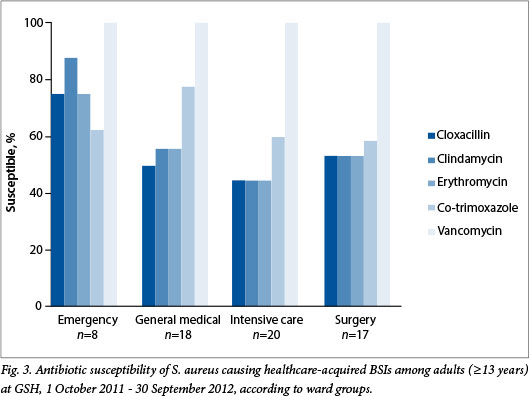
Resistance was higher among healthcare-acquired pathogens. Just over half of S. aureus isolates were susceptible to cloxacillin and to clindamycin (52.4% and 55.6%, respectively) (Fig. 3). No vancomycin resistance was detected in the 41 enterococcal isolates. Among the Enterobacteriaceae, only 41.4% were susceptible to amoxicillin/ clavulanate, 64.6% to gentamicin and 70% to ciprofloxacin (Fig. 3). ESBL production occurred in 39.6%. More than 99% were susceptible to the carbapenems, including ertapenem, meropenem and imipenem. Susceptibility to amikacin was 85.4%. Susceptibility to piperacillin/tazobactam was not reported owing to incomplete testing as a result of limitations of the Vitek 2 Gram-negative susceptibility card in use at the time (Product Correction/Field Safety Notice Vitek® 2 Piperacillin/Tazobactam Memo, communication from bioMerieux, 5 April 2011).
Pseudomonas aeruginosa showed relatively low susceptibility rates to most agents tested, ranging from 44.4% for imipenem and ciprofloxacin to 72.2% for ceftazidime (data not shown). Colistin was the only agent that retained high levels of susceptibility (91.7%). Susceptibility rates for piperacillin/tazobactam could not be reported for P. aeruginosa isolates for the same reason as above. A. baumanii complex also demonstrated low levels of susceptibility: only tobramycin and colistin retained susceptibility rates above 70% (72.1% and 96.6%, respectively) (data not shown).
Ward stratification
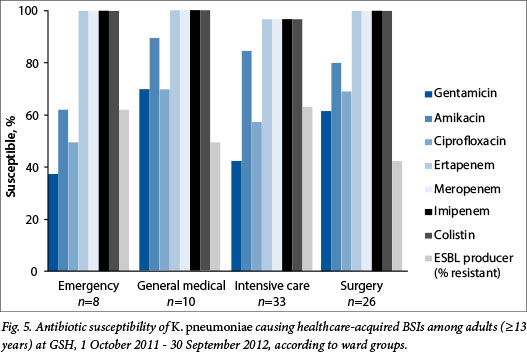
The ward distribution of healthcare-acquired organisms is shown in Table 3. Half (n=271, 49.9%) of all healthcare-acquired isolates were associated with ICUs. Of these, nearly a quarter were identified as A. baumanii complex (n=65, 24.0%), while just over one-tenth were K. pneumoniae (n=33, 12%). In contrast, 28% of isolates were associated with general surgical wards (n=153, 28.2%), with equal numbers of E. coli and K. pneumoniae predominating (n=26, 17.0%). Fewer healthcare-associated isolates were associated with emergency (n=34, 6.3%) and general medical wards (n=76, 14.0%), but among these, the predominant organisms were S. aureus (emergency n=8, 23.5%; general medical n=18, 23.7%), K. pneumoniae (emergency n=8, 23.5%; general medical n=10, 13.2%) and E. coli (emergency n=7, 20.6%; general medical n=11, 14.5%) (Table 3). The number of healthcare-acquired isolates associated with the maternity wards was very small (n=9), so no further analysis of these isolates is presented.
The susceptibility profiles of selected pathogens isolated from different wards are shown in Figs 4 - 6. There were no statistically significant differences in the proportions susceptible to antibiotics in different wards, apart from E. coli isolates in the emergency ward (n=7), which were significantly more susceptible (100%) to ciprofloxacin than isolates from the ICUs (40%, n=10), and Acinetobacter isolates in the ICUs (n=65), which were significantly less susceptible to tobramycin (67.7%) and to amikacin (39%) than those associated with surgical wards (n=14, 100% and 78% susceptibility, respectively) (p<0.05 for all comparisons).


Discussion
Three-quarters of BSIs in adults at GSH are healthcare acquired. Although this may be partially expected at a tertiary hospital receiving many patients referred from other institutions, and therefore fewer community-acquired infections, this high proportion does illustrate the potential for reduction in numbers of BSIs through preventive measures, e.g. prompt removal of unnecessary intravenous lines and urinary catheters, and through improved hand hygiene.
The predominance of Gram-positive organisms among community-acquired infections and Gram-negative organisms among healthcare-acquired infections was confirmed in this study. Overall the most prevalent organisms responsible for community-acquired (S. pneumoniae, E. coli, S. aureus and K. pneumoniae) and healthcare-acquired (Acinetobacter spp., Klebsiella spp., S. aureus, E. coli, enterococci, Enterobacter spp., P. aeruginosa, and coagulase-negative staphylococci) BSIs in our study were generally consistent with other recent studies.[18,19] Klebsiella spp., S. aureus and E. coli were the commonest causes of healthcare-acquired infections in all areas outside the ICUs, whereas Acinetobacter was common in the ICUs and rare in all other areas.
Community-acquired bloodstream pathogens in this hospital remain largely susceptible to the first-line antibiotics, but antibiotic resistance is relatively common among healthcare-acquired bloodstream pathogens. Given that more than 50% of S. aureus isolates are resistant to cloxacillin, empirical vancomycin is appropriate for serious healthcare-acquired S. aureus infections. Likewise, with 40% of healthcare-acquired Enterobacteriacae being ESBL producers, the use of ertapenem for suspected Gram-negative sepsis in this hospital seems justified. Amikacin is one possible alternative (85% susceptibility), but unfortunately it was not possible to investigate the proportion of isolates susceptible to the previously used combination of piperacillin-tazobactam and amikacin. In the ICUs, colistin or tobramycin may be needed for Acinetobacter infections. These conclusions are in line with contemporary hospital antibiotic recommendations.
Compared with a previous report that included data from GSH from 2010,[10] a lower proportion of healthcare-acquired S. aureus were susceptible to cloxacillin (52.4% v. 69%) and clindamycin (55.6% v. 71%) in the present study. The proportion of susceptible A. baumanii isolates was also lower in the present study with regard to ceftazidime (20.7% v. 49%), imipenem (18.4% v. 26%), meropenem (18.6% v. 23%) and gentamicin (39.1% v. 47%). The likely reasons for these differences are the stratification of isolates into community- and healthcare-acquired infections, as well as the exclusion of contaminants and duplicates.[10]
Our results confirm the low susceptibility rates for P. aeruginosa and A. baumannii that have been reported in SA national surveillance data and in studies from other countries (although numbers of isolates may be low for confident comparisons) and reinforce the reliance on colistin as the sole agent that retains activity against most strains.[20] The ward-stratified results suggest that the distribution of organisms and their susceptibility patterns do not differ significantly by ward, apart from the greater proportion and more resistant Acinetobacter isolates associated with ICUs. This finding may be limited by the small numbers of isolates per ward.
Surveillance of antibiotic resistance, locally as well as regionally and globally, is important. Consistent application of standard definitions will assist in this ongoing endeavour. As has been pointed out previously,[10] laboratory-based surveillance depends on the culture submission practices of clinicians. Data should be maintained and extractable with an emphasis on facilitating regular analysis and reporting. Reporting should be based on standards that can be widely accepted and applied.
Study limitations
The chief limitation of this study was our inability to retrospectively assess the accuracy of the contaminant classification. These may be genuine contaminants, but could also include isolates for which clinical information was lacking or represent instances of error or omission by the reporting microbiologist. The majority of the group designated as contaminants were coagulase-negative staphyloccci or corynebacteria, which are recognised skin flora. However, some fungi and mycobacteria were not classified; these are likely to be mostly community acquired. Small numbers of Enterobacteriaceae (n=34) and S. aureus (n=24) were probably incorrectly excluded from the analysis of pathogens; however, the number of these relative to the numbers included was low (290 and 93 included, respectively). Improvements to the reporting system, such as inclusion of additional reporting items to differentiate unclassified isolates, as well as monthly review of data, could minimise these deficiencies.
Another important limitation concerns the reporting of all ESBL producers as resistant to all cephalosporins. Potentially, a proportion of these isolates would be susceptible to cephalosporins, particularly cefepime. However, evidence suggests that carriage of multiple ESBL genes is common in SA,[21] and at the time of writing this matter is still under discussion by national microbiology groups. As the study spanned 2011 and 2012, the CLSI interpretive criteria that were applied differed over time. However, the changes were relatively few in number, and did not affect the major findings in relevant antibiotic susceptibilities in this study or alter the empirical treatment recommendations.
This report does not allow us to discuss trends in resistance patterns over time, as the analysis was conducted cross-sectionally. Additionally, the data do not allow us to comment on incidence of infection, either healthcare or community acquired. Changes in culture submission practices over time may have an impact on these reported susceptibility results. As with other studies of this nature, if clinicians are more likely to submit cultures for infections that are not responding to initial empirical treatment, the overall resistance rate will be inflated. We are not aware of any hospital-based outbreaks occurring during the study period that would have biased our results.
Conclusion
The distinction between community- and healthcare-acquired acquisition of infection is relevant and important to empirical treatment options of BSIs. The findings presented here suggest that healthcare-acquired bloodstream pathogens carry significant resistance phenotypes. As hospital antibiotic use places substantial selection pressure on circulating organisms, efforts must continue to be directed towards improving the appropriateness of antibiotic use. This includes selection of an effective antimicrobial agent, as well as prescribing the optimal dose and duration, for all clinically important bacterial infections.
Community-acquired BSIs in this region retain considerable susceptibility to the antibiotics commonly used to treat them. Antibiotic stewardship efforts will be supported by the selection of relatively narrow-spectrum antibiotics for community-acquired infections, even in the tertiary setting. Resistance should be monitored closely so that treatment and antibiotic stewardship practice can continue to be informed by local susceptibility patterns.
Differentiation of community- and healthcare-acquired BSIs provides additional useful information, and could be readily incorporated into routine laboratory practice by any laboratory that conducts similar clinical liaison, although measures to ensure accuracy of classification should be included.
Acknowledgements. The authors wish to acknowledge the National Health Laboratory Service of South Africa for providing the data to conduct this analysis.
References
1. Laupland KB. Defining the epidemiology of bloodstream infections: The 'gold standard' ofpopulation-based assessment. Epidemiol Infect 2012;141(10):2149-2157. [http://dx.doi.org/10.1017/S0950268812002725] [ Links ]
2. Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med 2005;352(1):39-47. [http://dx.doi.org/10.1056/NEJMoa040275] [ Links ]
3. Skogberg K, Lyytikainen O, Ollgren J, Nuorti JP, Ruutu P. Population-based burden of bloodstream infections in Finland. Clin Microbiol Infect 2012;18(6):E170-E176. [http://dx.doi.org/10.1111/j.1469-069L2012.03845.x] [ Links ]
4. Diekema DJ, Beekmann SE, Chapin KC, Morel KA, Munson E, Doern GV. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol 2003;41(8):3655-3660. [http://dx.doi.org/10.1128/JCM.4L8.3655-3660.2003] [ Links ]
5. Aiken AM, Mturi N, Njuguna P, et al. Risk and causes of paediatric hospital-acquired bacteraemia in Kilifi District Hospital, Kenya: A prospective cohort study. Lancet 2011;378(9808):2021-2027. [http://dx.doi.org/10.1016/S0140-6736(11)61622-X] [ Links ]
6. Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect Dis 2010;10(6):417-432. [http://clx.doi.org/10.1016/S1473-3099(10)70072-4] [ Links ]
7. Friedman ND, Kaye KS, Stout JE, et al. Health care-associated bloodstream infections in adults: A reason to change the accepted definition of community-acquired infections. Ann Intern Med 2002;137(10):791-797. [http://dx.doi.org/10.7326/0003-4819-137-10-200211190-00007] [ Links ]
8. French GL. Clinical impact and relevance of antibiotic resistance. Advanced Drug Deliv Rev 2005;57(10):1514-1527. [http://dx.doi.org/10.1016/j.addr.2005.04.005] [ Links ]
9. Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Clin Infect Dis 2006;42(Suppl 2):S82-S89. [ Links ]
10. Bamford C, Bonorchis K, Elliott E, et al. Antimicrobial susceptibility patterns of selected bacteraemic isolates from South African public sector hospitals, 2010. Southern African Journal of Epidemiologyand Infection 2011;26(4):243-250. [ Links ]
11. Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med 2004;10(12s):S122-S129. [http://dx.doi.org/10.1038/nm1145] [ Links ]
12. Grundmann H, Klugman KP, Walsh T, et al. A framework for global surveillance of antibiotic resistance. Drug Resist Updat 2011;14(2):79-87. [http://dx.doi.org/10.1016/j.drup.2011.02.007] [ Links ]
13. Bamford C, Goodway J, Hoffmann R Rapid identification and susceptibility testing of Gram-negative bacilli from blood cultures using the Vitek Southern African Journal of Epidemiology and Infection 2010;25(3):28-31. [ Links ]
14. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. CLSI document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute, 2012. [ Links ]
15. Clinical and Laboratory Standards Institute. Performance standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI document M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute, 2012. [ Links ]
16. Clinical and Laboratory Standards Institute. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Approved Guideline. 3rd ed. CLSI document M139-A3. Wayne, PA: Clinical and Laboratory Standards Institute, 2009. [ Links ]
17. Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev 2006;19(4):788-802. [http://dx.doi.org/10.1128/CMR.00062-05] [ Links ]
18. Brink A, Feldman C, Duse A, et al. Guideline for the management of nosocomial infections in South Africa. S Afr Med J 2006;96(7):642-652. [ Links ]
19. De Bus L, Coessens G, Boelens J, Claeys G, Decruyenaere J, Depuydt P. Microbial etiology and antimicrobial resistance in healthcare-associated versus community-acquired and hospital-acquired bloodstream infection in a tertiary care hospital. Diagn Microbiol Infect Dis 2013;77(4):341-345. [http://dx.doi.org/10.1016/j.diagmicrobio.2013.08.009] [ Links ]
20. Visser Kift E, Maartens G, Bamford C. Systematic review of the evidence for rational dosing of colistin. S Afr Med J 2014;104(3):183-186. [http://dx.doi.org/10.7196/SAMJ.7011] [ Links ]
21. Perovic O, Singh-Moodley A, Duse A, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010 - 2012. S Afr Med J 2014;104(8):563-568. [http://dx.doi.org/10.7196/SAMJ.7617] [ Links ]
 Correspondence:
Correspondence:
C Bamford
colleen.bamford@nhls.ac.za
Accepted 18 March 2015














