Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
SAMJ: South African Medical Journal
versión On-line ISSN 2078-5135
versión impresa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.105 no.1 Pretoria ene. 2015
http://dx.doi.org/10.7196/SAMJ.8814
FORUM
GENETICS IN MEDICINE
No evidence for clinical utility in investigating the connexin genes GJB2, GJB6 and GJA1 in non-syndromic hearing loss in black Africans
A WonkamI, V ; J BoschII, V; J J N NoubiapVI; K LebekoIII, V; N MakubaloIV, V; C DandaraV
IMedical geneticist trained at the University of Geneva, Switzerland, whose research interest is reflected in more than 70 publications with a focus on monogenetic diseases of people of African descent. He was the principal investigator on this project.
IIJason Bosch completed his MSc in human genetics on the project, with molecular analysis of the GJB2 and GJA1 genes
IIIDid all the experiments on the GJB6 gene for her honours degree in human genetics.
IVPaediatrician, spent two years in the medical genetics unit and did all the clinical phenotyping of the cohort from the Eastern Cape, South Africa, where she is currently practising.
VA molecular geneticist, supervised all the molecular experiments in the project. All these authors were affiliated to the Division of Human Genetics, Department of Clinical Laboratory Sciences, Faculty of Health Sciences, University of Cape Town, South Africa, during the course ofthe project.
VIJean Jacques Noubiap, ofthe Faculty of Medicine and Biomedical Sciences, University ofYaoundé I, Yaoundé, Cameroon, is a general practitioner with strong research interests. He performed all the clinical phenotyping of the Cameroonian cohort for his MD degree.
ABSTRACT
BACKGROUND: Deafness is the most common sensory disability in the world. Globally, mutations in GJB2 (connexin 26) have been shown to play a major role in non-syndromic deafness. Two other connexin genes, GJB6 (connexin 30) and GJA1 (connexin 43), have been implicated in hearing loss, but these genes have seldom been investigated in black Africans. We aimed to validate the utility of testing for GJB2, GJB6 and GJA1 in an African context.
METHODS: Two hundred and five patients with non-syndromic deafness from Cameroon and South Africa had the full coding regions of GJB2 sequenced. Subsequently, a carefully selected subset of 100 patients was further sequenced for GJB6 and GJA1 using Sanger cycle sequencing. In addition, the large-scale GJB6-D3S1830 deletion was investigated.
RESULTS: No pathogenic mutations that could explain the hearing loss were detected in GJB2, GJB6 or GJA1, and the GJB6-D3S1830 deletion was not detected. There were no statistically significant differences in genomic variations in these genes between patients and controls. A comprehensive literature review supported these findings.
CONCLUSION: Mutations in GJB2, GJB6 and GJA1 are not a major cause of non-syndromic deafness in black Africans and should not be investigated routinely in clinical practice.
Deafness is one of the leading causes of disability in the world. The prevalence of hearing loss is highest in South Asia and sub-Saharan Africa, attributable to poor healthcare systems where complications at birth as well as infections result in loss of hearing in the newborn.!1'
Screening for hearing loss in newborns is standard practice in many countries and is the most effective way of detecting hearing problems and reducing the negative effects of hearing loss. South Africa (SA) does not have a national screening programme. At best, only 7.5% of public hospitals provide screening for hearing loss, with only 1% providing universal screening.[2] The situation is even worse in the rest of sub-Saharan Africa.[3] Clinical presentation of hearing loss is extremely heterogeneous, ranging from mild to total hearing loss and presenting either as a single symptom or as one of many clinical features. The causes of hearing loss can be genetic or environmental. A recent review of the aetiology of childhood hearing loss showed that 48.3% of cases of hearing loss were of unknown cause, 30.4% were genetic and 19.2% were acquired.[4] In developing communities, the environment contributes significantly more to congenital hearing loss than in the developed world. In Africa, bacterial meningitis contributes to cases of hearing loss in infants and young children.[3] With improved healthcare there will be a reduction in cases of hearing loss caused by disease and an increase in the proportion attributable to genetics, the majority of which are non-syndromic.[1]
To date, 65 different genes, with many different causative mutations, have been identified that contribute to non-syndromic deafness.[5] Mutations in gap junction (GJ) genes, specifically GJB2 (connexin 26), have been shown to be the major contributors to deafness globally.[6]
GJ proteins (connexins) regulate functions of the cochlea
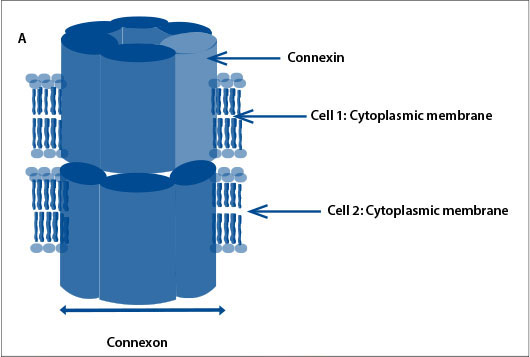
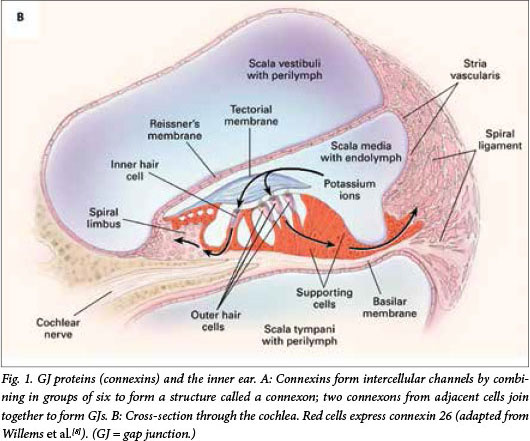
GJs are intercellular channels that allow ions, second messengers and small metabolites to be exchanged by adjacent cells. Connexins form intercellular channels by combining in groups of six to form a structure called a connexon.[7] Connexons from adjacent cells join together to form GJs (Fig. 1, A). The connexin proteins are named according to their weight; connexin 26 is a protein with a molecular weight of 26 kDa. The two major groups of connexins are the alpha and beta connexins, based on sequence similarity of the cytoplasmic loop. GJB2 was the second beta connexin gene to be identified. GJB2 is located on chromosome 13 and codes for the GJ protein connexin 26. Connexin 26 is involved in the transport of potassium ions and other small molecules and is expressed in the cochlea (Fig. 1, B).[8] It is believed that mutations in GJB2 (connexin 26 gene) affect its ability to transport potassium ions and therefore regulate the endocochlear potential required for nerve impulses involved in hearing, but the mode of action is not definitively elucidated.[9]
No mutations in connexin genes GJB2, GJB6 or GJA1 among Cameroonians and Xhosa South Africans
With the exception of Ghana,[10] mutations in GJB2 (connexin 26) have not been shown to be a major contributor to deafness in sub-Sahara Africa.[6] Could other potential candidate genes, GJB6 (connexin 30) and GJA1 (connexin 43), lead to non-syndromic deafness in Africans? The GJB6-D13S1830 deletion is present in up to 9.7% of people of European descent,[11] and represents the second leading genetic cause of non-syndromic deafness. When GJA1 mutations were detected in African Americans,[12]GJA1 emerged as a possible candidate for hearing loss in indigenous Africans. However, failure by investigators to differentiate between GJA1 and its pseudogene led to this hypothesis being discarded.[13]
We performed a series of molecular investigations and reviewed the literature with the aim of validating the clinical utility of testing for GJB2, GJB6 and GJA1 in the African context. As part of this validation, we recruited a total of 205 patients affected with non-syndromic hearing loss from a well-described Cameroonian cohort[3] and newly recruited black South Africans of Xhosa ancestry, the majority (85%) of whom had sensorineural deafness. All 205 patients were investigated for GJB2 gene, as previously reported.[14] A subset of 100 selected patients, with deafness likely to be of genetic cause (mostly familial cases) and who did not have any mutation in GJB2 gene, were investigated for mutations in the GJB6 and GJA1 genes.[15]
All the coding regions of GJB2, GJB6 and GJA1 were amplified and detection of del(GJB6-D13S1830) was also investigated.[14,15] In the GJB2 gene, two likely pathogenic mutations were detected in two unrelated Cameroonian participants, g.3741_3743delTTC (p.F142del) and g.3816G>A (p.V167M) in a single individual each and in the heterozygous state (Table 1). No pathogenic mutation was detected among the SA patients.[14] Phylogeny analysis of the sequence data from the Cameroonian and SA controls, together with that of various populations extracted from the 1000 Genomes Project, shows as expected that the SA patients and Cameroonian controls grouped with the other African populations. There was a low variance when comparing sequences in GJB2 in Africans with that of other population groups: the principal component analysis explains only 40% of the variations.[14] Specific sequence variants in the GJB2 gene in Africans could therefore not explain the low occurrence of mutations associated with non-syndromic deafness in this population.
In the GJB6 gene, none of the patients had the GJB6-D13S1830 deletion. Only one variant (rs145762940) was detected, in the heterozygous state, in the coding region of GJB6, leading to the c.480G>A (p.G160=) change.[15] Equally, in the GJA1, five variants were detected; one of these occurred in the intron, but none were known to be pathogenic.[15]
Few mutations in GJB2, GJB6 or GJA1 genes among other populations of African ancestry
Our report and review of the literature confirmed that GJB2 gene is of little significance in non-syndromic hearing loss in populations of African descent (Table 1).[10,14,16-20] In addition, by combining data from previously unstudied deaf Xhosa patients in SA, Cameroonian patients, previous studies in Africans and the 1000 Genomes Project, the analysis further supported the limited contribution of GJB2 genes in non-syndromic hearing loss in Africans. Interestingly, we reported two cases of keratitis-ichthyosis-deafness (KID) syndrome in two Cameroonian patients,[21] caused by mutations in GJB2. In both cases the mutation found (p.Asp50Asn) was the most common in many populations globally.[5,22] Adding to the established founder effect of the GJB2 mutations reported in European and Asian populations,[5] the data indicate that the high frequency of GJB2 mutations in non-syndromic hearing loss have evolved in Eurasian populations after their migration out of Africa, and spread with population migrations. Finally, at the genetic level, the Cameroonian population diversity mimics that of various ethnolinguistic groups in African populations;[23] it is anticipated that results from a carefully selected sample in this population could capture those of many other populations on the African continent.
As in previous studies in Africans,[17] African Americans and Caribbean Hispanics with GJB6 mutations,[19] we did not find either the GJB6-D13S1830 deletion or coding region changes. Similarly, no pathogenic variants were detected in GJA1, suggesting their non-implication in hearing loss among the Cameroonians and black South Africans studied,[15] as has been reported in African Americans.[13]
Clinical implications and research perspectives
From our analysis, there is no evidence that mutations in GJB2, GJB6 or GJA1 are associated with non-syndromic deafness in sub-Saharan African patients. We therefore recommend against routine use of either gene for clinical testing in patients of African ancestry. We suggest that future research should take advantage of the power of massively parallel sequencing to screen multiple genes at once. This approach has previously been shown to offer the best chance of uncovering the genetic causes of deafness in settings with a genetically diverse populations.[24]
Funding. The research was funded by the University of Yaoundé for clinical phenotyping and DNA extraction and by the National Health Laboratory Service, SA, for recruitment of SA samples, DNA extraction and molecular analysis. Jason Bosch was supported by funding provided by the University of Cape Town through the Ada and Bertie Levenstein Scholarship. Kamogelo Lebeko is supported by an internship funding provided by the South African National Research Foundation.
Acknowledgements. We thank Efata School for the Deaf and Blind for their assistance, and the patients and their families for their participation.
1. Stevens G, Flaxman S, Brunskill E, Mascarenhas M, Mathers CD, Finucane M; Global Burden of Disease Hearing Loss Expert Group. Global and regional hearing impairment prevalence: An analysis of 42 studies in 29 countries. Eur J Public Health 2013;23(1):146-152. [http://dx.doi.org/10.1093/eurpub/ckr176] [ Links ]
2. Swanepoel D1, Störbeck C, Friedland P. Early hearing detection and intervention in South Africa. Int J Pediatr Otorhinolaryngol 2009;73(6):783-786. [http://dx.doi.org/10.1016/j.ijporl.2009.01.007] [ Links ]
3. Wonkam A, Noubiap JJN, Djomou F, Fieggen K, Njock R, Toure GB. Aetiology of childhood hearing loss in Cameroon (sub-Saharan Africa). Eur J Med Genet 2013;56(1):1-6. [http://dx.doi.org/10.1016/j.ejmg.2012.09.010] [ Links ]
4. Korver AM, Admiraal RJ, Kant SG, et al. Causes of permanent childhood hearing impairment. Laryngoscope 2011;121(2):409-416. [http://dx.doi.org/10.1002/lary.21377] [ Links ]
5. Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. http://hereditaryhearingloss.org/ (accessed 3 May 2012). [ Links ]
6. Chan DK, Chang KW. GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 2014;124(2):E34-E53. [http://dx.doi.org/10.1002/lary.24332] [ Links ]
7. Mese G, Richard G, White TW. Gap junctions: Basic structure and function. J Invest Dermatol 2007;127(11):2516-2524. [http://dx.doi.org/doi:10.1038/sj.jid.5700770] [ Links ]
8. Willems PJ. Genetic causes of hearing loss. N Engl J Med 2000;342(15):1101-1109. [http://dx.doi.org/10.1056/NEJM200004133421506] [ Links ]
9. Iossa S, Marciano E, Franzé A. GJB2 gene mutations in syndromic skin diseases with sensorineural hearing loss. Curr Genomics 2011;12(7):475-785. [http://dx.doi.org/10.2174/138920211797904098] [ Links ]
10. Hamelmann C, Amedofu GK, Albrecht K, et al. Pattern of connexin 26 (GJB2) mutations causing sensorineural hearing impairment in Ghana. Hum Mutat 2001;18(1):84-85. [http://dx.doi.org/10.1002/humu.1155] [ Links ]
11. Del Castillo I, Villamar M, Moreno-Pelayo MA, et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med 2002;346(4):243-249. [http://dx.doi.org/10.1056/NEJMoa012052] [ Links ]
12. Liu XZ1, Xia XJ, Adams J, et al. Mutations in GJA1 (connexin 43) are associated with non-syndromic autosomal recessive deafness. Hum Mol Genet 2001;10(25):2945-2951. [http://dx.doi.org/10.1093/hmg/10.25.2945] [ Links ]
13. Paznekas WA, Boyadjiev SA, Shapiro RE, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet 2003;72(2):408-418. [http://dx.doi.org/10.1086/346090] [ Links ]
14. Bosch J, Noubiap JJN, Dandara C, et al. Sequencing of GJB2 in Cameroonians and black South Africans and comparison to 100 Genomes Project data support the need to revise strategy for discovery of nonsyndromic deafness genes in Africans. OMICS 2014;18(11):705-710. [http://dx.doi.org/10.1089/omi.2014.0063] [ Links ]
15. Bosch J, Lebeko K, Nziale JJ, Dandara C, Makubalo N, Wonkam A. In search of genetic markers for nonsyndromic deafness in Africa: A study in Cameroonians and black South Africans with the GJB6 and GJA1 candidate genes. OMICS 2014;18(7):481-485. [http://dx.doi.org/10.1089/omi.2013.0166]. [ Links ]
16. Trotta L, Iacona E, Primignani P, et al. GJB2 and MTRNR1 contributions in children with hearing impairment from Northern Cameroon. Int J Audiol 2011:50(2):133-138. [http://dx.doi.org/10.3109/14992027.2010.537377] [ Links ]
17. Kabahuma RI, Ouyang X, Du LL, et al. Absence of GJB2 gene mutations, the GJB6 deletion (GJB6-D13S1830) and four common mitochondrial mutations in nonsyndromic genetic hearing loss in a South African population. Int J Pediatr Otorhinolaryngol 2011:75(5):611-617. [http://dx.doi.org/10.1016/j.ijporl.2011.01.029] [ Links ]
18. Gasmelseed NMA, Schmidt M, Magzoub MMA, et al. Low frequency of deafness-associated GJB2 variants in Kenya and Sudan and novel GJB2 variants. Hum Mutat 2004:23(2):206-207. [http://dx.doi.org/10.1002/humu.9216] [ Links ]
19. Samanich J, Lowes C, Burk R, et al. Mutations in GJB2, GJB6, and mitochondrial DNA are rare in African American and Caribbean Hispanic individuals with hearing impairment. Am J Med Genet A 2007;143A(8):830-838. [http://dx.doi.org/10.1002/ajmga.31668] [ Links ]
20. Pandya A, Arnos KS, Xia XJ, et al. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med 2003:5(4):295-303. [http://dx.doi.org/10.1097/01.GIM.0000078026.01140.68] [ Links ]
21. Wonkam A, Noubiap JJN, Bosch J, Dandara C, Toure GB. Heterozygous p.Asp50Asn mutation in the GJB2 gene in two Cameroonian patients with keratitis-ichthyosis-deafness (KID) syndrome. BMC Med Genet 2013:14(1):81-87. [http://dx.doi.org/10.1186/1471-2350-14-81] [ Links ]
22. Gasparini P, Rabionet R, Barbujani G, et al. High carrier frequency of the 35delG deafness mutation in European populations. Eur J Hum Genet 2000:8(1):19-23. [http://dx.doi.org/10.1038/sj.ejhg.5200406] [ Links ]
23. Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science 2009:324(5930):1035-1044. [http://dx.doi.org/10.1126/science.1172257] [ Links ]
24. Shearer AE, Black-Ziegelbein EA, Hildebrand MS, et al. Advancing genetic testing for deafness with genomic technology. J Med Genet 2013:50(9):627-634. [http://dx.doi.org/10.1136/jmedgenet-2013-101749] [ Links ]
 Correspondence:
Correspondence:
A Wonkam
ambroise.wonkam@uct.ac.za
Accepted 13 October 2014.
This month in the SAMJ...

Colleen Aldous* is a senior lecturer in the School of Clinical Medicine at the University of KwaZulu-Natal, Durban. She is a medical scientist who holds a PhD in science education. She is involved in mentorship of research at the postgraduate level across several medical disciplines, e.g. surgery, orthopaedics, dermatology, paediatrics, opthalmology, general medicine and psychology. Her own research interest is within the discipline of human genetics, specifically in the epidemiology thereof. She is currently part of the National Steering Committee and Working Group for the review of the National Department of Health Policy Guidelines for Human Genetics Services.
*Aldous CM, Adhikari M, Rout CC. The research component of specialist registration - a question of alligators and swamps? A personal view. S Afr Med J 2015:105(1):21-22. [http://dx.doi.org/10.7196/SAMJ.8732]

David Lee Skinner* is a surgical consultant in the intensive care unit at King Edward VIII Hospital, Durban.
He is an Honorary Clinical Fellow and member of the Perioperative Research Group in the Department of Anaesthetics and Critical Care, University of KwaZulu-Natal. He completed his MMed in acute kidney injury (AKI) in trauma patients and has research interests in AKI, blunt thoracic trauma and perioperative medicine. Further research goals involve the investigation and management of AKI associated with crush injury. He believes that trauma and critical care research are essential for improving rational service delivery in our resource-constrained healthcare system.
*Skinner DL, den Hollander D, Laing GL, Rodseth RN, Muckart DJJ. Severe blunt thoracic trauma: Differences between adults and children in a level I trauma centre. S Afr Med J 2015:105(1):47-51. [http://dx.doi.org/10.7196/SAMJ.8499]

Prof. Mike Levin* is head of the Division of Asthma and Allergy in the Department of Paediatrics, University of Cape Town and Red Cross War Memorial Children's Hospital, Cape Town. He lectures on allergy to medical students and professionals, presents at national and international allergy congresses, and has a special interest in the epidemiology of allergy in Africa, severe allergies, anaphylaxis and patient support and education.
*Levin ME, Gray CI, Goddard E, et al., for the South African Food Allergy Working Group (SAFAWG). South African food allergy consensus document 2014. S Afr Med J 2015:105(1):62-65. [http://dx.doi.org/10.7196/SAMJ.9098]