Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
SAMJ: South African Medical Journal
versão On-line ISSN 2078-5135
versão impressa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.103 no.12 Pretoria Dez. 2013
MOLECULAR GENETICS
STR null alleles complicate parentage testing in South Africa
A B Lane
BSc (Hons), MSc, PhD, Division of Human Genetics, School of Pathology, National Health Laboratory Service, and Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
ABSTRACT
BACKGROUND: Null alleles complicate parentage testing because they do not contribute positively to phenotypes.
OBJECTIVES: To survey South African populations for null alleles at short tandem repeat (STR) loci used in parentage testing.
METHODS: Paternity case data were scanned for apparent contradictions compared with Mendelian inheritance that could be due to null alleles. Estimates of null allele frequencies were obtained from tallies of apparent carriers.
RESULTS: Three of 15 loci appeared to have null alleles at appreciable frequencies and five showed no evidence of null alleles. A null allele at the vWA locus reached a frequency of ~0.3% in the black population and approximately three times this frequency in the coloured population. No apparent vWA null carriers were detected in whites suggesting that the Khoisan were the major contributors of the null alleles to coloureds. The apparent genotypes of a sample of TPOX null carriers changed from homozygous to heterozygous when they were retyped using different polymerase chain reaction primers. The revealed allele was allele 6 in every case. A D13S317 null allele was detected at relatively low frequencies in the black and coloured samples but 2/145 Indian (Asian) parents appear to be carriers, suggesting that it could be common in Indians.
CONCLUSION: Three of the 15 forensically relevant STR loci investigated had null alleles at significant frequencies in South African populations. Failure to allow for the presence of null alleles can have a large impact on the outcome of parentage tests.
Although the scientific principles of paternity testing were established towards the middle of the 20th century[1-4] using serogenetic markers such as blood groups, they apply equally now that DNA analysis[5] has become the method of choice. When they are efficient, parentage tests 'exclude' the vast majority of wrongly accused individuals by revealing inconsistencies with normal Mendelian inheritance between the genotypes of alleged parent and child. At present, most laboratories carry out paternity testing by identifying the alleles present at sets of unlinked autosomal short tandem repeats (STRs) containing loci (microsatellites). The usual procedure used for genotyping involves polymerase chain reaction (PCR) amplification followed by electrophoresis in a genetic analyser. Commercially available kits[5] that were primarily developed for analysing crime-scene material are widely used by parentage test laboratories to facilitate the profiling process.
STRs are ideal for matching crime-scene DNA to that of criminals and victims, but their relatively high mutation rates complicate their use in parentage and kinship testing. The possibility of inheriting an allele that mutated during sex-cell formation means that an alleged parent, who does not share an allele with their alleged child at every tested locus, cannot be excluded with absolute certainty. In addition, the use of a limited number of STR loci (kits usually enable the typing of 9 - 15 autosomal STR loci) can very occasionally result in failure to exclude a wrongly accused person who may, by chance, carry copies of all the alleles that the child inherited from its true parent. For these reasons, the conclusions arrived at in disputed parentage cases reflect the likelihood of parentage and are not absolute.
The weight of evidence favouring parentage is usually assessed by contrasting two likelihoods.[6] The first is the (genetic) probability of observing the genotypes (or more correctly, phenotypes) of the tested individuals on condition that they are a biological family, and the second is the probability of observing the same genotypes on condition that someone other than the alleged parent is the child's true parent. The number obtained by dividing the first likelihood by the second, the likelihood ratio (LR), is also called the parentage index. Because unlinked loci (more or less independent systems) are used, the LR obtained from the data on one locus can be multiplied by that from the next and so on to produce a combined likelihood ratio (CLR). A CLR of 1 (i.e. 1/1) indicates that the chance of parentage is equal to the chance of non-parentage. Any CLR that is less than unity is evidence of non-parentage, and any CLR that is greater is evidence of parentage. If a parentage test yields a large CLR, e.g. one that is >100 or 1 000, then there is little doubt that the alleged parent is in fact the biological parent. Alternatively, if a parentage test yields a very small CLR, for instance one that is less than 0.01 or 0.001, then there is little doubt that the alleged parent is not the biological parent.
When using kits to type >9 STR loci, cases with CLRs that are between 0.01 and 100 usually exhibit one or occasionally two inconsistencies with normal Mendelian inheritance and the results should be viewed as inconclusive. A possible reason for an inconclusive CLR is that a mutated allele was transmitted to the child. Another is that a tested alleged parent has copies of most, but not all of the child's alleles because he/she is a close relative of the true parent. The lowering effects of these possibilities on the CLR are compounded in cases where either the undisputed parent or the alleged parent is unavailable for testing (deficiency cases). A third reason for an inconclusive CLR can be the transmission of a null allele from parent to child. This possibility should be considered in all cases where the alleged parent and the child appear to be homozygous for different alleles. Failure to allow for the possible segregation of a null allele can make a huge difference to the evidence of parentage and may result in a report that supports an incorrect conclusion. The information required for calculating an LR when a null allele could have been inherited is the frequencies of the overt alleles of the individuals concerned, the null allele frequency in the population and the mutation rate of the STR locus. All of the above information, except the null allele frequency, is usually available; however, no published estimates of null allele frequencies in South African populations have been made to date. As a first step towards remedying this, a survey was performed on a database of paternity test results with the aim of arriving at ball-park estimates of null allele frequencies at 15 of the 20 or so commonly used STR loci[7] in the major South African populations. In addition, the effects of ignoring the possible presence of null alleles on the strength of parentage evidence is also demonstrated using case work data.
Methods
Estimating null allele frequencies
Routine parentage tests were carried out with the aid of either the AmplSTR Profiler, COfiler, Profiler Plus or Identifiler (Applied Biosystems) or the Powerplex 16 (Promega) kits. The results were scanned for inconsistencies of the type encountered when null alleles are segregating. This was done by querying the database (locus by locus) for mother-child or alleged father-child pairs, where the child and parent (or alleged parent) appeared to be homozygous. Such cases were then checked to see whether the overt alleles were different and whether this was the sole inconsistency, i.e. the alleged parent was not 'excluded' by results obtained at other loci in the profiles. The total number of 'non-excluded' males and mothers were separately obtained through the use of the SQL 'COUNT' function and added together (N in Table 1). An example of the type of inconsistency looked for is shown in Fig. 1. Here, a mother appears to be homozygous for a different allele to her apparently homozygous child. This pattern suggests that a null allele is segregating; particularly as a mutation that could have altered a 16 repeat allele to a 14 repeat allele (a two repeat change) is a rare event.[8]
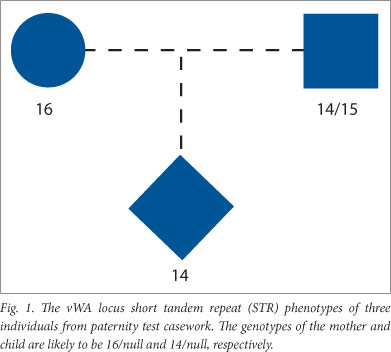
All apparently homozygous undisputed mothers who had a different STR phenotype to their apparently homozygous children were counted as probable null allele carriers. The same criteria were used with alleged fathers, where only one of the 9 or 15 loci tested showed an apparent inconsistency of the type shown in Fig. 1.
A sample of the apparent TPOX null allele carriers who were identified in the way described above were re-profiled using the COfiler or Identifiler kits, which use different primers to those employed in the Powerplex 16 kit.
Ball-park null allele frequencies were estimated from the number of apparent null allele carrying 'parents' and the total number of (non-excluded) parents in the database after correcting for ascertainment bias. In single-child paternity cases, ascertainment bias was compensated for by counting every apparent null allele carrier parent as two null carriers. The reason for this is that only half of the null carriers with one child who came for paternity testing were expected to have passed on their null allele (the other half would have passed on their normal allele). According to binomial distribution theory, 25% of null carriers with two tested children will fail to transmit a null allele to either child (50% will transmit their null to one of their two children and 25% will transmit it to both). This means that 1 extra carrier should be added for every 3 two-child carriers encountered and by extension, 1 extra null carrier should be added for every 7 three-child carrier parent, etc.
For those loci where no evidence of null alleles was found, a conservative working null allele frequency could be calculated by pretending that the next parent to be encountered would be a null allele carrier. To counter ascertainment bias, this 'null allele carrier' should be taken to be 2 null allele carriers. Finally, this number (2) is divided by: the number of parents in the database + 2, to give a conservative estimated null allele frequency.
Compliance with Hardy-Weinberg expectation was checked on an earlier dataset which also contained 'excluded' alleged fathers, by the exact method[9] using Arlequin software.[10]
The effect of allowing for the possible presence of a null allele
Paternity test trios (mother, child and alleged father) in which a vWA null allele appeared to be segregating were selected. An initial CLR was calculated from the phenotypes of the apparent null carrier and child on the assumption that mutation alone could have been responsible for the inconsistency. The estimated mutation rates used were those given in the STRbase (http://www.cstl.nist.gov/div831/ strbase/mutation.htm) and the model used for LR calculation in the face of a possible mutational event was that proposed by Charles Brenner (http://dna-view.com). Two-repeat (two-step) mutations were taken to be 10 times less common than single-step mutations, three-step mutations were taken to be 10 times less common than two-step mutations, etc. A second CLR was then calculated by taking into account both the possible presence of a null allele (at the locus specific frequency estimated from the data in Table 1) and the possibility of mutation.
A software programme (available on request) was written for the purpose of calculating LRs in cases where null alleles could be segregating.
Results
Estimating null allele frequencies
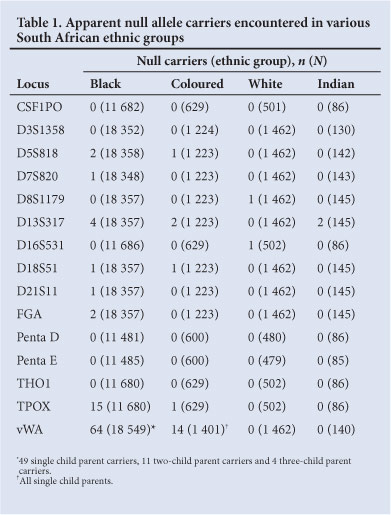
Table 1 shows the apparent null allele carriers encountered in the four main South African ethnic groups (the total numbers of non-excluded 'parents' are given in parentheses). No null/null individuals were observed within the entire sample.
The numbers of individuals typed for all of the above loci are not equal because the kits used initially catered for fewer loci than those used later, and because the Promega and Applied Biosystems kits do not facilitate the typing of identical sets of loci. The proportions of the different ethnic groups who were tested also varied over time. Only 3/15 loci (D13S317, vWA and TPOX) appear to have appreciable null allele frequencies and 5 (CSF1PO, D3S1358, Penta D, Penta E and THO1) showed no evidence of null alleles at all. The presence of null alleles was not sufficient to distort 'genotype' frequencies significantly from Hardy-Weinberg expectation; e.g. the p-values obtained for the D13S317, vWA and TPOX data sets from the black group were 0.471, 0.100 and 0.492, respectively. It is not surprising that no null/null allele homozygotes were observed; the expected homozygote frequency of the vWA null allele in the black and coloured populations was 1/104 058 and 1/10 203, respectively; whereas the sample sizes from these two groups were only 18 549 and 1 401, respectively.
The vWA null allele
In the black population: 49 single-child null carrier parents, 11 two-child carrier parents and 4 three-child carrier parents were found. To correct for ascertainment bias, 49 null carriers were added for the 49 single-child carrier parents, 2.75 carriers to the 11 two-child carrier parents observed and 0.5714 carriers to the 4 three-child carrier parents. This gave a theoretical total of 116.32 vWA null allele carriers among 18 549 parents, which translates to an estimated null allele frequency of 0.00314.
There were 14 apparent vWA null allele carriers (all with one tested child) among 1 401 coloured parents. After correcting for ascertainment bias, the estimated frequency of this allele is 0.0099, which makes it approximately 3.2 times more common in coloureds than in blacks. No apparent vWA null carriers were detected in the white or Indian groups. All of the carriers in the remaining systems were single-child parents.
Distribution of the vWA null allele in South African ethnic groups
Assignments of ethnic group are likely to be biologically imprecise because many of the subjects were from large towns and cities where liaisons between members of different ethnic groups are common. In addition, the ethnic groups of children recorded by their mothers usually correspond to the mother's self-declared group regardless of the alleged father's. Walton[11] showed that the vWA null allele encountered in local populations represents a deletion of part of the vWA STR, which can be detected by electrophoresis following PCR amplification using flanking primers. Shantikumar,[12] from the same laboratory, went on to show that this allele resulted from the deletion of 174 nucleotides of the vWF gene which encompasses the vWA STR. The data suggest that the vWA null allele frequency is different in different black ethnic groups (Fig. 2). The Nguni groups (Zulu, Xhosa and Swazi) seem to have higher frequencies than the Sotho-Tswana, Venda and Tsonga/Shangaan. A χ2 test (carried out after pooling two groups with low expected carrier numbers) to assess the possibility that the observed differences in the black groups are merely due to chance, indicated that this is extremely unlikely (χ2=24.568, degrees of freedom 7, p=0.0009). This indicates that an averaged null allele frequency should be used with caution in parentage calculations as different regions with different ethnic group representations may show different average vWA null allele frequencies.
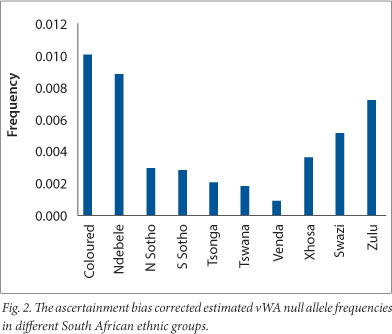
Coloureds show the highest vWA null allele frequency, which might seem surprising since no evidence of a vWA null allele was found in any of the 1 518 whites investigated. However, direct evidence (electrophoresis PCR products obtained using flanking primers) of a high frequency (~0.047) in the Khoisan was found by Walton,[11] which suggests that the Khoisan could have been the major donors of the allele to the coloureds. The allele also appears to have a high frequency in the Ndebele, which may have resulted from gene flow from the Khoisan, although genetic drift or sampling variation could also be responsible.
The TPOX null allele
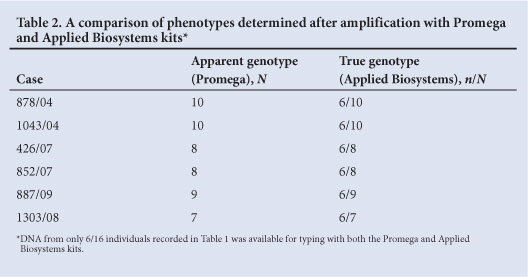
TPOX null alleles were only observed in samples from the black and coloured population. However, it is possible, although unlikely, that their apparent absence from the white and Indian groups is due to small sample sizes. The phenotypes of all apparent TPOX null allele carriers who were tested changed when they were retyped using an Applied Biosystems kit (Table 2). Each was apparently homozygous when typed with the Promega kit and heterozygous when typed with an Applied Biosystems kit. In addition, the uncovered or revealed allele was allele 6 in every case, which means that a subset of allele 6 in the black and coloured populations is linked to the primer binding site variant.
The D13S317 null allele
A D13S317 null allele was detected at relatively low frequencies in samples from the black and coloured groups. However, 2/145 Indian (Asian) parents appeared to be carriers, suggesting that the null allele could be common in Indians.
Making allowance for the possible presence of a vWA null allele in paternity cases
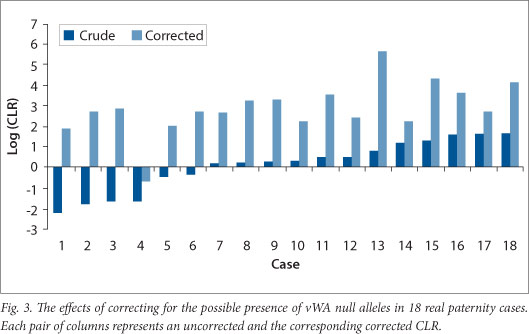
The effect of making allowance for the possible presence of a vWA null allele in cases where the alleged parent and child appeared homozygous for different alleles was fairly dramatic. This was done using the overall null allele frequency estimated for the 'black' population. The CLR increased in every case when the possible presence of a null allele was considered in addition to the possibility of mutation (Fig. 3); the smallest increase was 8 fold and the largest 442 770 fold. The latter's CLR (Case 13 in Fig. 3) changed from 7 to 442 770.
Discussion
In the context of parentage testing, Gjertson et al.,[6] state that, A null allele is an allele that does not contribute to the phenotypic result of the test'. When PCR products are the phenotype, failure of amplification from a target DNA strand represents the presence of a null allele. Genetic variation which results in mismatches between PCR primers and their targets is the most common cause of STR null alleles.[6] The data presented above indicate that the local TPOX null allele incorporates a primer binding variant that is in linkage disequilibrium with a 6-repeat allele. Sun et al., as cited in STRBase, reported the loss of allele 11 in some samples typed with the Promega's Powerplex 16 kit, which were correctly detected using the Applied Biosystems Identifiler kit. This variant is clearly different to the local TPOX null allele where allele 6 is 'lost'.
The most frequently encountered null allele in the present study represents a deletion of part of the vWA locus, which includes the downstream primer binding sites of both the Applied Biosystems and Promega kits. This allele's frequency appears to be higher in the coloured population than in blacks. As no evidence of its presence was found in whites, it seems likely that this allele has largely flowed into the coloured population from the Khoisan. The vWA null allele could either have entered the black population's gene pool during a long period of interbreeding with the Khoisan or it could have been present in the progenitor of both the Khoisan and black groups. The latter seems more likely as it is found in all the major South African black ethnic groups. The vWA null allele appears to be more common in Nguni language speakers, particularly the Xhosa. This is in accordance with other evidence of extensive gene flow between the Khoisan and Nguni.[13] The apparently high vWA null allele frequency in the Ndebele may be the result of sampling error (only 342 individuals sampled), genetic drift or it may have resulted from admixture with the Khoisan. Drabek et al.[14] found nine apparent vWA null carriers in a sample of 212 African Americans while evaluating a novel set of 'miniplex' primers. These are clearly different to the local vWA null allele because they turned out to be due to primer binding variants in either allele 12, 13 or 14 and all their carriers had corresponding heterozygous phenotypes when typed with the Applied Biosystems Identifiler and the Promega Powerplex 16 kits.
It must be admitted that the method used for estimating null allele frequencies is crude. It was assumed that every parent who appeared to be homozygous for a different allele compared with his/her apparently homozygous child was a null allele carrier. In some rare instances the child's phenotype could have resulted from a mutational event in a homozygous parent and the transmission of a copy of the same allele to the child by its other parent. Another consideration is that some of the alleged fathers who were assumed to be carriers may not have been biological fathers but rather homozygous individuals who happened to share an allele with the child at all but one of the tested loci. Both possibilities are believed to be so rare that they are unlikely to make a significant impact on the estimated frequencies.
It can be seen from Fig. 3 that ignoring the possible presence of the vWA allele may lead to incorrect conclusions regarding parentage. Six of the 18 cases studied had CLRs (calculated by taking mutation into account), which were <1 suggesting non-parentage. When the possible presence of a null allele in addition to mutation was taken into account, 5/6 had CLRs >1 and all 5 had CLRs >100.
Conclusion
Evidence of three relatively common null alleles was found in the South African black and coloured populations. One, at the vWA locus, represents a deletion; the second, at the TPOX locus, is due to variation within a primer-binding site; and the nature of the third, at the D13S317 locus, is unknown at present. The existence of these null alleles should be taken into account when DNA STR data are being interpreted in parentage and kinship investigations.
References
1. Essen-Möller E. Die Beweiskraft derÄhnlichkeit im Vaterschaftsnachweis-theoretische Grundlagen. Mitt Anthrop Ges (Wien) 1938;68:9-53. [ Links ]
2. Essen-Möller E, Quensel C. Zur Theorie des Vaterschafsnachweises auf Grund vonÄhnlichkeitsbefunden. Z Ges Gerichtl Med 1939;31:70-96. [ Links ]
3. Ihm P. Die Mathematischen Grundlagen, vor allem für die statistische Auswertung des serologischen und anthropologischen Gutachtens. In: Hummel K, ed. Die medizinische Vaterschafts Begutachtung mit Biostatistische m Beweis. Stuttgart: Fischer, 1961:128-145. [ Links ]
4. Gürtler H. Principles of blood-group statistical evaluation of paternity cases at the University Institute of Forensic Medicine Copenhagen. Acta Med Leg Soc (Liege) 1956;9:83-93. [ Links ]
5. Butler JM. Fundamentals of Forensic DNA Typing. San Diego: Elsevier Academic Press, 2010:147-173. [ Links ]
6. Gjertson DW, Brenner CH, Baur MP, et al. ISFG: Recommendations on biostatistics in paternity testing. Forensic Sci Int: Genetics 2007;1(3-4):223-231. [http://dx.doi.org/10.1016/j.fsigen.2007.06.006] [ Links ]
7. Butler JM. Genetics and genomics of core STR loci used in human identity testing. J Forensic Sci 2006;51:253-265. [ Links ]
8. Dauber E-M, Kratzer A, Neuhuber F, et al. Germline mutations of STR-alleles include multi-step mutations as defined by sequencing of repeat and flanking regions. Forensic Sci Int: Genetics 2012;6(3):381-386. [http://dx.doi.org/10.1016/j.fsigen.2011.07.015] [ Links ]
9. Guo S, Thompson E. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992;48(2):361-372. [http://dx.doi.org/10.2307/2532296] [ Links ]
10. Excoffier L, Laval L, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online 2005;1:47-50. [ Links ]
11. Walton A. Characterisation of a vWA null allele in the South African population. BSc (Hons) in Human Genetics. Research Report 2004. Faculty of Science, University of the Witwatersrand, Johannesburg, South Africa. [ Links ]
12. Shantikumar K. The common vWA null allele in South African populations. BSc (Hons) in Human Genetics. Research Report 2005. Faculty of Science, University of the Witwatersrand, Johannesburg, South Africa. [ Links ]
13. Nurse GT, Weiner JS, Jenkins T. The Peoples of Southern Africa and Their Affinities. Oxford: Clarendon Press, 1985:219-228. [ Links ]
14. Drabek J, Chung DT, Butler JM, McCord BR. Concordance study between miniplex STR assays and a commercial STR typing kit. J Forensic Sci 2004;49(4):859-860. [ Links ]
 Correspondence:
Correspondence:
A B Lane
(tony.lane@mweb.co.za)
Accepted 12 August 2013.














