Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
SAMJ: South African Medical Journal
versión On-line ISSN 2078-5135
versión impresa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.103 no.8 Pretoria ene. 2013
RESEARCH
MammaPrint Pre-screen Algorithm (MPA) reduces chemotherapy in patients with early-stage breast cancer
K A GrantI; C WrightII; J P ApffelstaedtIII; E MyburghIII, V; R PienaarVI; M de KlerkVII; M J KotzeII
INat Dip Med Tech, NHD, MTech; Department of Biomedical Sciences, Faculty of Health and Wellness, Cape Peninsula University of Technology, Cape Town, South Africa. Division of Anatomical Pathology, Department of Pathology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
IINat Dip Med Tech, MB ChB, MMed, FCPath, FRCPath, FIAC, PhD; Division of Anatomical Pathology, Department of Pathology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa. National Health Laboratory Service, Port Elizabeth, South Africa
IIIDr Med, MMed (Surg), FCS (SA), MBA. Department of Surgery, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
IVMB ChB, FCS (SA), MMed (Surg). Department of Surgery, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa. VPanorama Medi-Clinic, Cape Town, South Africa
VMB ChB, MMed, RadT. GVI Oncology, Panorama Medi-Clinic, Cape Town, South Africa
VIMB ChB, MFam Med, MBA, DCH. Metropolitan Health Group and University of Stellenbosch Business School, Cape Town, South Africa
VIIBSc, BSc (Hons), MSc, PhD. Division of Anatomical Pathology, Department of Pathology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
ABSTRACT
BACKGROUND: Clinical and pathological parameters may overestimate the need for chemotherapy in patients with early-stage breast cancer. More accurate determination of the risk of distant recurrence is now possible with use of genetic tests, such as the 70-gene MammaPrint profile.
OBJECTIVES: A health technology assessment performed by a medical insurer in 2009 introduced a set of test eligibility criteria - the MammaPrint Pre-screen Algorithm (MPA) - applied in this study to determine the clinical usefulness of a pathology-supported genetic testing strategy, aimed at the reduction of healthcare costs.
METHODS: An implementation study was designed to take advantage of the fact that the 70-gene profile excludes analysis of hormone receptor and human epidermal growth factor receptor 2 (HER2) status, which form part of the MPA based partly on immunohistochemistry routinely performed in all breast cancer patients. The study population consisted of 104 South African women with early-stage breast carcinoma referred for MammaPrint. For the MammaPrint test, RNA was extracted from 60 fresh tumours (in 58 patients) and 46 formalin-fixed, paraffin-embedded (FFPE) tissue samples.
RESULTS: When applying the MPA for selection of patients eligible for MammaPrint testing, 95 of the 104 patients qualified. In this subgroup 62% (59/95) were classified as low risk. Similar distribution patterns for risk classification were obtained for RNA extracted from fresh tumours v. FFPE tissue samples.
CONCLUSIONS: The 70-gene profile classifies approximately 40% of early-stage breast cancer patients as low-risk compared with 15% using conventional criteria. In comparison, more than 60% were shown to be low risk with use of the MPA validated in this study as an appropriate strategy to prevent chemotherapy overtreatment in patients with early-stage breast cancer.
Breast cancer is the most common cause of cancer mortality in women worldwide and places an increasing burden on health services in both the developing and developed world.[1] Although mortality has decreased over the past two decades, incidence rates continue to increase, particularly in developing countries where the majority of cases are diagnosed at an advanced stage. Breast cancer survival rates also vary greatly between high- and low-income countries, with low survival rates explained mainly by late detection in resource-restricted countries.
Breast cancer is a heterogeneous disease characterised by genetically distinct subtypes that differ in their response to treatment.[2] In approximately 20% of breast tumours, over-expression of the human epidermal growth factor receptor 2 (HER2) gene is associated with a poor prognosis and resistance to tamoxifen and methotrexate-containing chemotherapy regimens, while targeted immunotherapy with Herceptin (trastuzumab) reduces the recurrence rate in these patients by about 50%. Patients with hormone-dependent breast cancer usually respond to a 5-year course of selective oestrogen receptor (ER) modulators, ovarian suppression or aromatase inhibitors. Chemotherapy is generally the only treatment option available for patients with the most aggressive subtype, known as basal-like or triple-negative breast cancer. The benefits of chemotherapy for triple-negative and HER2-positive tumours are well documented. Conversely, only a small minority of patients with ER-positive, progesterone receptor (PR) positive and HER2-negative tumours will benefit from chemotherapy, while all patients with such tumours offered chemotherapy are exposed to its side-effects.
Traditional guidelines for the use of adjuvant chemotherapy use tumour extent, lymphatic involvement and cellular morphology as a surrogate for tumour biology. Unfortunately, this correlates poorly and tends to overestimate the risk of systemic disease. Over the past 20 years, extensive research has been performed into genetic profiling of breast tumours to more accurately identify poor prognostic tumour subtypes. Several tests have been developed commercially, of which MammaPrint and Oncotype Dx are available in South Africa (SA).
MammaPrint uses a highly versatile microarray platform reflects the hallmarks of cancer-related biology for classification of early-stage breast cancer into low- or high-risk groups for chemotherapy selection. [3] It has been validated in at least 3 independent studies, with clinical utility confirmed by a recent prospective 5-year follow-up trial.[4] Further refinement of the test in 2010 enabled sub-classification of tumour tissue into luminal A, luminal B, HER2-positive and basal-like subtypes (BluePrint). This distinction cannot be achieved by standard pathology[5] and demonstrates the power of the microarray platform as a discovery tool for ongoing research, based on the ~25 000 genes in the human genome evaluated during development of the MammaPrint test.
In a separate readout from the same microarray platform, quantitative gene expression levels of ER, PR and HER2 (TargetPrint) are determined and provide additional confirmation of immunohistochemistry (IHC) and/or fluorescence in situ hybridisation (FISH) results at the RNA level. High concordance with IHC/FISH, demonstrated in the first 800 primary breast cancers of patients enrolled in the Microarray In Node-negative and 1 to 3 positive lymph node Disease may Avoid ChemoTherapy (MINDACT) trial and in over 600 samples previously tested by Roepman et al.[6] for ER, PR and HER2 status, confirmed the robustness of the microarray platform. Multi-gene expression tests that are based on real-time polymerase chain reaction (RT-PCR) methodology, such as Oncotype Dx, have been criticised for lack of reproducibility in determination of HER2 overexpression, which forms an integral part of the recurrence score.[7,81]
MammaPrint is the only test cleared by the US Food and Drug Administration (FDA), and this independent confirmation of analytical and clinical validation provides a high level of confidence in test performance.
A critical step during the implementation phase of new technologies is to understand where any additional information provided by a genomic test could fit into the context of the current clinicopathological prognostication of early-stage breast carcinoma. Traditionally, tumours with ER and/or PR overexpression that are HER2-negative, have shown the lowest benefit to the addition of adjuvant chemotherapy. The addition of anthracycline chemotherapy to Herceptin is considered standard of care, and HER2-positive tumours were expected to benefit little by further genetic profiling. Due to the lack of alternative treatment options in ER-, PR- and HER2-negative tumours, chemotherapy remains the only adjuvant option, and the use of MammaPrint would not alter the treatment plan. Therefore, the ability to base clinical decision-making on microarray analysis after exclusion of triple-negative and HER2-positive patients following standard IHC and FISH assessments, was an important consideration.
The likelihood that it would be most cost-effective to use MammaPrint in such a clinical intermediate group (ER-positive and HER2-negative) in the resource-poor SA context, prompted a local medical insurer to subject the 70-gene MammaPrint test to a health technology assessment (HTA) in 2009. The results of the HTA indicated a break-even-point for cost-effectiveness of the test (at R22 000 per test) at approximately R88 000 for the cost of chemotherapy.'91 In this model, conventional criteria for chemotherapy treatment, namely the St Gallen Index and Adjuvant! Online, were replaced with a newly defined MammaPrint Pre-screen Algorithm (MPA), which is further evaluated in the present study.
Methods
Study design
This study was designed to evaluate the appropriateness of a screening step for the selection of patients for MammaPrint, where the test would have the greatest impact to reduce chemotherapy, namely in hormone receptor-positive, HER2-negative early-stage breast cancer patients. The MPA took advantage of the fact that the 70-gene profile excludes assessment of ER, PR and HER2, already determined routinely in all breast cancer patients, using less expensive IHC and FISH tests. In a separate readout of the microarray platform, the ER, PR and HER2 status was assessed quantitatively as a confirmation of these IHC results, and to help resolve cases with equivocal results.
The study protocol was approved by the Ethics Research Committee of the University of Stellenbosch (ref. number N09/06/166).
Study participants
The study population included 104 SA patients with early-stage breast cancer referred for the MammaPrint test from participating clinicians. Two of these patients presented with >1 tumour. An online questionnaire was used to document clinical characteristics. Additional information was obtained from patient records for comparative analysis.
Laboratory analysis
Histopathological diagnosis, including nodal status and assessment of ER, PR and HER2 status, was performed using standard IHC techniques and established pathology protocols. FISH was used to determine HER2 status in all 2+ immunohistochemistry-equivocal cases and to confirm gene amplification in 3+ IHC-positive cases. In samples taken from the same tumour with differing results, performed at different laboratories, 1+ IHC-negative cases may also have been subjected to FISH.
For the MammaPrint test, tumour samples were obtained from fresh surgical biopsies (n=60) or formalin-fixed, paraffin-embedded (FFPE) tissue (n=46). Samples were transported to Agendia in the Netherlands under an export permit obtained from the SA Department of Health. Microarray analysis was performed only on specimens with at least 30% tumour as evaluated by an experienced histopathologist, in accordance with FDA requirements.
Defining the MammaPrint criteria
The international MammaPrint selection criteria, that allow inclusion of ER-negative and HER2-positive breast cancer, were modified for use in the HTA being evaluated in this study (Table 1).
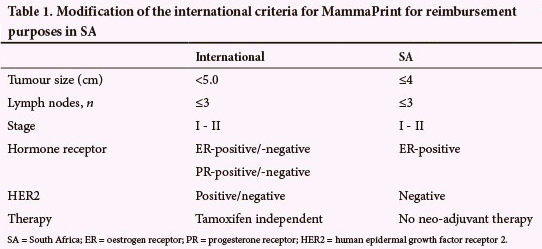
Statistical analysis
A cross tabulation with the Chi-square test was performed to compare the distribution of MammaPrint low- v. high-risk profiles obtained between fresh tumour biopsies and FFPE tissue. A one-way analysis of variance (ANOVA) was conducted to test for a relationship between risk classification and age of breast cancer diagnosis at the time that the MammaPrint test was performed.
Results
Table 2 summarises the clinical characteristics of the study population - 104 female early-stage breast cancer patients. Of the 106 tumour specimens analysed with MammaPrint, 60% were classified as low risk and 40% as high risk for breast cancer recurrence. Similar distribution patterns for MammaPrint low- v. high-risk profiles were obtained, irrespective of whether fresh tumour biopsies or FFPE tissue was used. The mean histopathological tumour size was 14.6 mm.
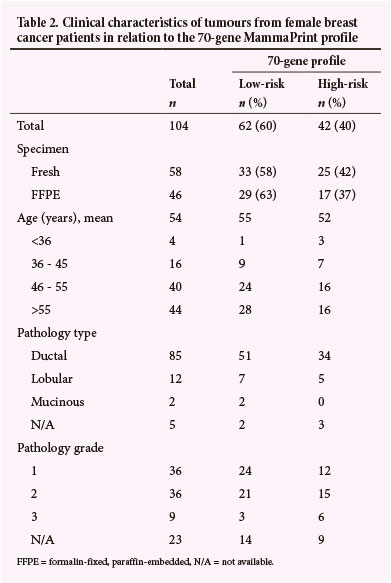
No statistically significant relationship was found between risk classification and age of the patient at diagnosis (p=0.19), although younger patients tended to have a high-risk MammaPrint profile. The youngest patient (24 years old) had a high-risk tumour and the oldest (78 years old) had a low-risk gene profile. Two patients, both older than 55 years of age, had multicentric lobular carcinomas. One patient was identified as low risk for both tumours, while the second patient had both a high- and low-risk tumour according to the MammaPrint profile, and was documented as high risk in Table 1. The tumours of these 2 patients were not graded. IHC results for ER, PR and HER2 status are not shown in Table 1 as these assessments formed part of the selection criteria for MammaPrint using the MPA.
When applying the criteria for selection of SA patients eligible for MammaPrint testing (Table 1), 95/104 patients qualified. In this subgroup 59 (62%) were classified as low risk and 36 (38%) as high risk, based on the 70-gene profile. The remaining 9 patients were referred for MammaPrint before implementation of the MPA in 2009 or opted for gene profiling outside the approved algorithm for medical aid reimbursement, as they refused chemotherapy based on clinicopathological features alone.
Discussion
The MPA developed in SA to reduce healthcare costs was validated in this translational research study as an appropriate strategy for selection of chemotherapy in patients with early-stage breast cancer. The process used to introduce the 70-gene MammaPrint test into the SA healthcare system included modification of the test selection criteria for local use, based on: (i) clinical experience, and (ii) establishment of an online database tool to provide support for reimbursement by funders and to facilitate long-term health outcome studies.
A total of 104 early-stage breast cancer patients were included in the present study. When we applied the MPA for determination of MammaPrint testing eligibility, 95 patients qualified for gene profiling. Using the MammaPrint test, 62% of patients in this clinical intermediate subgroup were re-classified as low-risk for recurrence. The HTA, using a model that replaced the conventional criteria for chemotherapy treatment (i.e. the St Gallen Index and Adjuvant! Online) with the MammaPrint risk profile, initially indicated a theoretical increase from 15% to >40% of patients who could be spared adjuvant chemotherapy. The fact that HER2-positive breast cancer accounts for approximately 20% of all breast cancers and that these cases, as well as ER-negative tumours on IHC, are excluded from genomic testing when applying the SA criteria, could explain the relatively high percentage of low-risk tumours in our study cohort.
Early-stage breast cancer patients with low-risk tumours can indeed safely be spared chemotherapy, as demonstrated recently by Drukker et al.[4] in the first prospective 5-year follow-up study performed for gene-profiling of breast cancer. One subject in this SA cohort with bilateral, MammaPrint low-risk tumours has remained disease free since 2008, despite a family history of early-onset breast cancer. The patient tested negative for mutations in the BRCA1 and 2 genes analysed separately using DNA extracted from blood. She was included in our clinical outcome study performed in the first 50 SA breast cancer patients,[10] which showed that 2 high-risk tumours and none of the low-risk tumours have recurred. This finding supported the treatment changes reported in 46% of cases discordant with conventional criteria, leading to a reduction in chemotherapy recommendations.
The proportion of low-risk patients identified using the MPA (62%) is in accordance with the results of Hartmann et al.,[11] who reported 38/60 (63%) female patients as low risk using similar eligibility criteria for the 70-gene MammaPrint test: pT1c-3, pN0-1a, grade 2/3, hormone receptor-positive, HER2-negative tumour. While previous studies suggested that the amount of prognostic information contained in 4 standard IHC assays - ER, PR, HER2, and Ki-67 - is similar to that of the Oncotype DX recurrence score,[12] Hartmann et al.[11] confirmed that MammaPrint provides further risk stratification over and above that obtained by standard pathology tests. These findings supported the development of the MPA as a pre-screen prior to MammaPrint testing in the resource-poor SA context. In a direct comparison of the cost-effectiveness of the 2 different genomic profiling tests using a Markov model, Yang et al.[13] demonstrated MammaPrint to be the most cost-effective. Several studies demonstrated greater cost-effectiveness in patients with ER-positive tumours, which is in accordance with the HTA performed in the SA population, based partly on the exclusion of patients with ER-negative and HER2-positive tumours for MammaPrint testing.
For breast cancer patients to benefit from new technologies, an increased focus on translational research is required to move basic science into clinical and public medical practice.[14] The first step to incorporate gene expression profiling into clinical management of early-stage breast cancer patients in SA involved a feasibility study of the surgical procedure for specimen collection for the MammaPrint test.[15] The 70-gene MammaPrint test initially required a fresh tumour biopsy taken during surgery for microarray analysis, which was placed into a preservative solution and shipped to the reference laboratory at room temperature. Since the beginning of 2012, the option of using FFPE tumour tissue has also become available for the MammaPrint test. Initially, using a fresh biopsy to perform the test, a relatively high failure rate of 23% (18/78) was experienced, mainly due to the inability to meet the FDA requirement of at least 30% tumour cell tissue. It also required the pre-operative planning for tissue collection and storage of the tissue in an appropriate medium until the final tumour histopathology became available. In comparison, all 46 specimens tested to date using FFPE have been successfully analysed using fixed tissue collected during surgery or core needle biopsies.
The use of FFPE, instead of fresh surgical biopsies, greatly enhanced the accessibility and convenience of the MammaPrint test in SA and elsewhere. Our finding of a similar distribution for MammaPrint low- v. high-risk profiles using fresh tumour biopsies or FFPE tissue confirmed the feasibility of RNA extraction from FFPE specimens for use in microarray analysis. Analytical validation using FFPE specimens was confirmed by excellent agreement with IHC/FISH results for the determination of HER2 status using microarray analysis and was found to be particularly useful to help resolve borderline cases (data not shown). Discrepancies may arise due to subjective interpretation of IHC and FISH results, arbitrary cut-off levels for positive results that may differ between the commercial kits, or due to a low percentage of invasive tumour in the tissue block when performing RT-PCR.[7,8] Our pathology-supported genetic testing strategy enables the evaluation of a combination of tests most likely to overcome the limitations of a single test procedure.
For both specimen types, the low-risk MammaPrint profile was shown to be approximately 60%, which implies safe avoidance of chemotherapy in this subgroup.
The HTA-based recommendations for use of the MammaPrint 70-gene profile in SA breast cancer patients initially included a requirement that the breast tumour should range between 1 and 4 cm in diameter. Recently, this requirement was modified to include tumours <1 cm and core biopsies, due to the change of sampling from fresh biopsies to FFPE specimens for microarray analysis. The international criteria for MammaPrint include tumours up to 5 cm and are independent of ER/HER2 status and treatment regimes. Furthermore, stage I or II disease, with a maximum of 3 nodes positive for disease, are eligible for MammaPrint referral. Although axillary lymph node status is an important prognostic factor, 25 - 30% of node-positive patients remain free of distant metastasis. Mook et al.[16] have demonstrated that the 70-gene signature can predict disease outcome in breast cancer patients with 1 - 3 positive lymph nodes. The conclusion was that MammaPrint accurately identifies patients with an excellent disease outcome in node-positive breast cancer, who may be safely spared adjuvant chemotherapy. Some patients in our cohort refused chemotherapy, yet opted to have gene profiling done outside the approved protocol for medical aid reimbursement. These included patients with 4 nodes positive for disease, triple-negative or HER2-positive tumours. Those found to be low risk despite a clinical high-risk profile could be reassured that chemotherapy might be safely avoided, as supported by the findings of Glück et al.[5] The results of this retrospective analysis on prospectively collected tumour specimens accurately predicted response to chemotherapy and showed a 5-year survival benefit in luminal B, HER2-positive and basal-type tumours.[5] The identification of a biological HER2-negative subgroup despite IHC/ FISH positivity may potentially lead to an extension of the MPA and further saving of healthcare costs in SA.
Use of gene profiling raises the confidence levels of clinicians regarding their treatment decisions.[9] Traditionally, results generated in the laboratory rely on the clinician to make a diagnosis and provide the appropriate treatment. Evaluation of the MPA in routine clinical practice performed in this study led to the development and implementation of an integrative pathology-supported genetic testing service for MammaPrint, whereby gene profiling is combined with pathological measurements to identify subgroups of patients requiring different treatment strategies. An online database tool is used to provide support for reimbursement by funders and to facilitate long-term health outcome studies. This integrative software tool, freely available to clinicians, should be seen as a platform where biology and the clinical components of medical judgement converge to assist clinicians in planning treatment of their patients.
The continuing rise in healthcare costs is unsustainable without changes in how cancer care is provided and reimbursed by funders. A process has therefore been initiated to develop a coverage policy for genomic tests by insurers in SA, based on the MammaPrint experience. Implementation of the MPA as a screening step for selection of patients for MammaPrint had a significant impact on reducing chemotherapy in early-stage breast cancer patients. The cost-saving implications of this approach support incorporation of the comprehensive microarray platform into treatment planning to (i) select chemotherapy in relevant early-stage breast cancer patients, (ii) confirm receptor status by providing quantitative gene expression assessment, as well as (iii) provide molecular subtyping of luminal A and B verifying receptor pathway activity.
Acknowledgments. We thank the many medical schemes, administrators and managed care organisations in SA and Namibia that reimbursed the MammaPrint test for breast cancer patients, without whom this study would not have been possible. Dr Zandile Dunn is acknowledged for her role in the development of a coverage policy. Dr Henry Davis is acknowledged for highlighting the need for appropriate a clinical protocol for application of MammaPrint in SA. Dr L Serfontein is acknowledged for defining the test selection criteria used for MammaPrint, together with EM and RP. This information was performed independently from the motivation provided by JPA requested for the health technology assessment. Dr K Baatjes is acknowledged for specimens collected at the Tygerberg Academic Hospital, as are Drs E Murray, D Pillay, H du Toit, C van Eeden, C Jacobs, C Coetzee, C-A Benn, G Philips, E Bouwer, D Coolen and C Struwig for patient referral from the private sector. Histopathologists Drs K Brundyn, G Swart and S Nayler are acknowledged for preparation of specimens used for MammaPrint. Drs L Heslop and M de Villiers are thanked for their contributions to the introduction of the MammaPrint service in SA through workshop presentations, and Prof. M Kidd for the statistical analysis. This work is part of a thesis to be submitted in fulfilment of the requirements for a PhD degree from Stellenbosch University.
Conflicts of interest. This work is based on the research supported in part by the National Research Foundation (grant no. 86417). The authors wish to thank the Cape Peninsula University of Technology (CPUT) research fund for providing financial support for the study. The Cancer Association of SA and CPUT Conference Committee are acknowledged for the travel grants applied to present the initial data at the 2013 St Gallen Congress in Switzerland. MJK is a director and shareholder of Gknowmix (Pty) Ltd that has developed a database tool for research translation under the auspices of the SA Medical Research Council. The Support Program for Industry Innovation is gratefully acknowledged for financial support towards development (2007 - 2012) of the GknowmixTM Genetic Knowledge Integration System and Clinical & Scientist Interface Model. These inventions firstly involved the development of a laboratory interface for Agendia in the Netherlands to provide SA patients access to the MammaPrint test. Secondly, a central online database was established using the variables initially defined by JPA, who also contributed to the HTA as requested by Metropolitan Health Risk Management.
References
1. Benson JR, Jatoi I. The global breast cancer burden. Future Oncol 2012;8(6):697-702. [http://dx.doi. org/10.2217/fon.12.61] [ Links ]
2. Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: How special are they? Mol Oncol 2010;4(3):192-208. [http://dx.doi.org/10.1016/j.molonc.2010.04.004] [ Links ]
3. Tian, S, Roepman P, van't Veer LJ, Bernards R, de Snoo F, Glas A. Biological functions of the genes in the MammaPrint breast cancer profile reflect the hallmarks of cancer. Biomarker Insights 2010;5:129-138. [http://dx.doi.org/10.4137/BMI.S6184] [ Links ]
4. Drukker CA, Bueno-de-Mesquita JM, Retèl VP, et al. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer 2013;133(4):929-936. [http://dx.doi.org/10.1002/ijc.28082] [ Links ]
5. Glück S, de Snoo F, Peeters J, Stork-Sloots L, Somlo G. Molecular subtyping of early-stage breast cancer identifies a group of patients who do not benefit from neoadjuvant chemotherapy. Breast Cancer Res Treat 2013;139(3):759-767. [http://dx.doi.org/10.1007/s10549-013-2572-4] [ Links ]
6. Roepman P, Horlings HM, Krijgsman O, et al. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res 2009;15(22):7003-7011. [http://dx.doi.org/10.1158/1078-0432.CCR-09-0449] [ Links ]
7. Dabbs DJ, Klein ME, Mohsin SK, Tubbs RR, Shuai Y, Bhargava R. High false negative rate of HER2 quantitative reverse transcription polymerase chain reaction of the Oncotype DX test: An independent quality assurance study. J Clin Oncol 2011;29(32):4279-4285. [http://dx.doi.org/10.1200/JCO.2011.34.7963] [ Links ]
8. Dvorak L, Dolan M, Fink J, Varghese L, Henriksen J, Gulbahce HE. Correlation between HER2 determined by fluorescence in situ hybridization and reverse transcription-polymerase chain reaction of the Oncotype DX Test. Appl Immunohistochem Mol Morphol 2013;21(3):196-199. [http://dx.doi.org/10.1097/PAI.0b013e3182632ff5] [ Links ]
9. Bateman C. Breast cancer breakthrough in gene profiling? S Afr Med J 2009;99(11):780-782. [ Links ]
10. Apffelstaedt JP, Kotze MJ. Five years of transcriptional profiling of breast tumours in a South African cohort. Abstract, EHPBA/ASSA SAGES 2011. Cape Town, South Africa 12 -16 April 2011. S Afr J Surg 2011;49(2):90-104. [ Links ]
11. Hartmann S, Gerber B, Elling D, Heintze K, Reimer T. The 70-gene signature as prognostic factor for elderly women with hormone receptor-positive, HER2-negative breast cancer. Breast Care 2012;7(1):19-24. [http://dx.doi.org/10.1159/000336552] [ Links ]
12. Cuzick J, Dowsett M, Pineda S, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29(32):4273-4278. [http://dx.doi.org/10.1200/JCO.2010.31.2835] [ Links ]
13. Yang M, Rajan S, Issa AM. Cost effectiveness of gene expression profiling for early stage breast cancer. Cancer 2012;118(20):5163-5170. [http://dx.doi.org/10.1002/cncr.27443] [ Links ]
14. Schully SD, Benedicto CB, Gillanders EM, Wang SS, Khoury MJ. Translational research in cancer genetics: The road less traveled. Public Health Genomics 2011;14:1-148.[http://dx.doi.org/10.1159/000272897] [ Links ]
15. Kotze MJ, Malan J, Pienaar R, Apffelstaedt J. The role of molecular genetic testing in modern breast health management. SA Fam Pract 2005;47(9):38-40. [ Links ]
16. Mook S, Schmidt MK, Viale G, et al. The 70-gene prognosis signature predicts outcome in breast cancer patients with 1-3 positive nodes in an independent validation study. Breast Cancer Res Treat 2009;116(2):295-302. [http://dx.doi.org/10.1007/s10549-008-0130-2] [ Links ]
 Correspondence: M J Kotze (maritha@sun.ac.za)
Correspondence: M J Kotze (maritha@sun.ac.za)














