Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
SAMJ: South African Medical Journal
versión On-line ISSN 2078-5135
versión impresa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.101 no.8 Pretoria ago. 2011
ORIGINAL ARTICLES
Part V. Surveillance activities
C Bamford; A Brink; N Govender; D A Lewis; O Perovic; M Botha; B Harris; K H Keddy; H Gelband; A G Duse
Keywords: surveillance; antibiotic (antimicrobial) resistance; acute respiratory infection; enteric infections; sexually transmitted infections
The critical importance of robust antimicrobial resistance (AMR) surveillance in South Africa cannot be overemphasised. Without knowing what the resistance situation is, it is impossible to develop appropriate antibiotic treatment guidelines and associated essential drug lists (EDLs) and to create and update evidence-based policies both at institutional and national levels. The broader benefits of AMR surveillance data include:
• Determining incidence rates of hospital-acquired infections (HAIs) and identifying the associated causative organisms and their AMR profile to feed into hospital guidelines and more appropriate treatment for infected patients. This in turn allows early interventions by infection prevention and control (IPC) so as to minimise further spread of AMR organisms.
• Profiling local or regional AMR patterns to inform selection of AMR screening practices in specific health care facilities (HCFs).
• Educating health care staff about the impact of AMR and about issues in antibiotic use and misuse.
• Monitoring trends over time to signal whether interventions are having the desired effect.
• Comparing South Africa with other countries in the region and around the world to facilitate sharing intervention experience.
South Africa has a good start at AMR surveillance, but it can and must be improved. For most AMR infections, surveillance data are laboratory and therefore organism centred, which limits the ability to differentiate between colonisation and infection with AMR organisms. It is also not possible to determine the clinical impact of AMR. A major shortcoming is that AMR surveillance is currently limited to a minority of HCFs, which does not reflect the extent of AMR across South Africa. The very limited profiling of AMR in the community needs to be addressed. Finally, the variability of surveillance methodology used makes it impossible to compare rates and trends across institutions.
The first part of this section describes studies that have identified serious AMR issues in South Africa which require urgent monitoring; these have provided compelling evidence of the need, and possible methods, for AMR surveillance.
AMR surveillance in South Africa
Surveillance of AMR in South Africa has in the past decade been carried out regularly by two main groups, with contributions from other parties. The involved groups are the National Antibiotic Surveillance Forum (NASF), currently known as the South African Society for Clinical Microbiology (SASCM), and the Group for Enteric, Respiratory and Meningeal disease Surveillance (GERMS). Additionally, the STI Reference Centre of the National Institute for Communicable Diseases (NICD), in collaboration with the National Department of Health (NDoH), performs sexually transmitted infection (STI) antibiotic resistance surveillance. In July 2010, a laboratory-based antimicrobial surveillance system was introduced by the Antimicrobial Resistance Reference Unit (AMRRU) of the NICD for HAI-associated Staphylococcus aureus and Klebsiella pneumoniae isolates collected from patients at designated sentinel sites throughout South Africa. Full characterisation of the resistance mechanisms of these isolates, as well as their molecular epidemiology, will be determined.
The National Antibiotic Surveillance Forum
The NASF was a voluntary professional organisation of medical microbiologists formed in 2002 with the key objective of monitoring AMR patterns in the public and private health sectors in South Africa. In 2009 NASF was superseded by the SASCM, which incorporated all surveillance activities, as well as involvement in other issues of concern to clinical microbiologists.
AMR surveillance in the public sector
In the public sector NASF/SASCM carries out retrospective laboratory-based surveillance of selected invasive pathogens isolated from blood and cerebrospinal fluid specimens at academic hospitals.
Methodology of the NASF/SASCM public sector AMR surveillance data system
NASF public sector surveillance relies on submission of data from the National Health Laboratory Service (NHLS) laboratories (Table I) that participate on a voluntary basis. The participating laboratories have been principally those serving academic tertiary care hospitals, although there has been some flux in the number of participating laboratories and in their catchment populations.
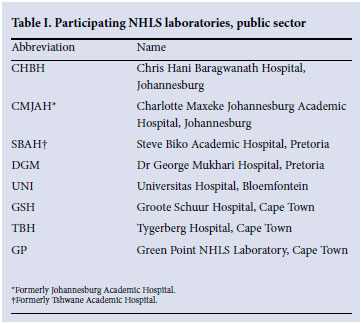
Laboratories submit AMR data on selected organisms isolated from blood cultures and cerebrospinal fluids by completing a standardised form. These organisms include: Streptococcus pneumoniae, Haemophilus influenzae, Neisseria meningitidis, Streptococcus group B, Enterococcus faecalis, S. aureus, Salmonella Typhi, non-typhoidal Salmonella, Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., Pseudomonas aeruginosa, Acinetobacter baumanii complex, Candida albicans spp. and Cryptococcus neoformans. Only blood culture and cerebrospinal fluid isolates are chosen since it can be assumed that, even in the absence of clinical information, isolates from these sites almost always represent clinically significant infections. The particulars of the pathogen- antibiotic combinations that are reported on are reviewed and updated regularly by a designated committee. All isolates are tested against a range of specified antibiotics.
All NASF surveillance data depend on the accurate identification and antimicrobial susceptibility testing (AST) performed at local laboratory level as no retesting is carried out at a central or reference laboratory. Different laboratories may use different methods for identification and AST and these methods may change over time. However, all the participating laboratories undertake regular internal and external quality assurance and many are accredited by the South African National Accreditation System (SANAS). Furthermore, all laboratories utilise Clinical and Laboratory Standards Institute (CLSI) criteria to perform and interpret antimicrobial susceptibilities, although different laboratories may implement annual updates of CLSI criteria at varying times.
Within the local laboratory, data are recorded either via software designed for the laboratory information system, or by review of various paper-based record-keeping systems. The available software is labour-intensive and not user-friendly, resulting in potential transcription errors. Extraction of minimum inhibitory concentration (MIC) values is particularly problematic and can be critical if changes in cut-offs complicate determination of temporal trends. Duplicate isolates are excluded to minimise bias due to over-representation of those patients whose cultures were performed most frequently. Only data on final, laboratory-authorised results are included. Monitoring of the quality of data submitted is achieved through self-reported answers to periodic questionnaires.
Data are collected at local level by a medical technologist or by a trainee microbiologist, and checked by an on-site pathologist before quarterly submission to a national co-ordinator. The national co-ordinator receives reports from the individual laboratories, and interrogates the data critically before collating an annual report. The report is reviewed by an editorial committee before dissemination via publications in local journals, or the organisation's website, or scientific presentations at meetings and conferences.
Strengths and limitations of the NASF/SASCM public sector surveillance system
The strengths of the NASF/SASCM public health surveillance system include:
• Because it is a nationwide programme it provides AMR data for all main regions in South Africa and allows for detection of similarities and differences between different areas.
• As it has been in existence for a number of years this allows for comparisons and determination of AMR trends.
• It addresses clinically relevant invasive pathogens and reports AMR to antibiotics that are generally available in the public sector.
• The large number of isolates for which AMR data are collected minimises the effects of errors or unusual patterns of resistance.
• Data are provided by generally competent laboratories. Important limitations include:
• Because this is a solely laboratory-based AMR surveillance system:
• there is no way to correlate patient outcomes with AMR data
• community-versus hospital-acquired infections cannot be differentiated
• the primary site of infection cannot always be determined
• submission of specimens for culture is dependent on clinicians whose test request practices may vary between different institutions.
• Uniformity is lacking with regard to data extraction methods.
• Surveillance is limited to large academic centres with an on-site microbiologist and AMR data from many smaller HCFs and rural areas are therefore lacking.
• Because participation is voluntary, time constraints experienced by participating members result in delays in submission of data.
To improve the NASF/SASCM public sector AMR surveillance system, the following resolutions taken at a workshop in September 2010 will be implemented in the next 6 - 12 months:
• Reinforce the importance of timely and consistent implementation of updated CLSI guidelines to facilitate standardisation between laboratories.
• Standardise AMR data collection procedures between the private and public sectors.
• Improve training in the use of computer-based epidemiology software programs, as well as in the interpretation of the data generated.
• Establish contacts with established surveillance programmes such as the European Antimicrobial Resistance Surveillance System (EARSS).
• Disseminate, through an editorial committee, the results of surveillance through brief, targeted and user-friendly reports.
• Investigate the possibility of obtaining surveillance data in terms of detailed information on individual isolates rather than through cumulative susceptibility results.
• Investigate the possibility of improving laboratory request forms to facilitate collection of clinical data.
• Establish centres of excellence for detection of emerging resistance.
Private sector AMR surveillance
Private surveillance data for various pathogens from various sources can be accessed on the website www.fidssa.co.za of the Federation of Infectious Diseases Societies of Southern Africa. AMR data from the private sector in South Africa are compiled from a laboratory information system, Meditech, which is used by all private laboratory groups and enables participating laboratories to extract standardised and reproducible AMR data and relevant parameters. Apart from this obvious advantage, similar limitations for the NASF/SASCM public sector AMR surveillance data pertain to the private sector AMR surveillance approach.
In the past, one private laboratory in Johannesburg, which participated in the SENTRY international antimicrobial surveillance programme, documented the prevalence of extended-spectrum β-lactamase (ESBL) production in Enterobacter cloacae and of oxacillin resistance in blood culture isolates of nosocomially acquired S. aureus among hospitalised patients in several Johannesburg private hospitals. As these results may not have been representative of the rest of private hospitals in South Africa, a wider study was prompted under the auspices of the NASF. It aimed to examine the susceptibility of important invasive Gram-negative pathogens and S. aureus in private health care institutions on a nationwide basis.1 Included was an investigation of the prevalence of ESBL production in selected Enterobacteriaceae cultured from all clinical specimens. All laboratories in private hospitals in South Africa's five largest cities participated.
The study clearly had several limitations and highlighted problems in the surveillance of pathogens isolated from patients in private hospitals. Susceptibility testing of the study isolates was not performed at a single site, nor was uniform methodology used. Furthermore, multidrug resistance (MDR) among invasive strains was not determined. Other limitations included the low numbers of isolates tested in some smaller centres and, more importantly, a lack of distinction between community-and hospital-acquired pathogens. Typical of laboratory-based surveillance, no clinical information was documented relating to colonisation or clinical significance, particularly in cases of ESBL-producing Gram-negative pathogens; this included the impact of resistance on outcome. Typing of ESBL-producing isolates was not performed. It is therefore uncertain whether cross-infection or clonal spread may have occurred to possibly account for the differences in ESBL rates in different localities. Additional problems highlighted in this study include the lack of standardisation in detection of glycopeptide resistance among isolates of S. aureus.
A second private national study, 'Emergence of extensive drug-resistance (XDR) among Gram-negative bacilli in South Africa - moving a step closer', was reported in 2008.2 It documented new developments, particularly with regard to increases in ESBL production as well as emergence of carbapenem resistance in invasive strains of K. pneumoniae, E. coli and Enterobacter spp. Once again strains were isolated from patients in private health care institutions, but from seven major centres in South Africa. The methods employed were similar to those described previously.1 The study was conducted from 1 July 2007 to 31 December 2007, and a total of 1 241 blood culture isolates were tested; E. coli (N=503) K. pneumoniae (N=548), and Enterobacter spp. (N=190).
The study highlighted:
• High levels of resistance to 'key workhorse' antibiotics used against Gram-negative pathogens in the health care institutions surveyed
• Significant prevalence of broad-spectrum antibiotic inactivating enzymes, in particular ESBLs in some centres, and other resistance mechanisms affecting fluoroquinolones and aminoglycosides in strains of invasive Enterobacteriaceae
• Considerable differences in the prevalence of resistance and ESBL production between the various cities
• The emergence of carbapenem resistance among the species in some centres.
These results emphasised the need for routine antimicrobial surveillance at least at regional level, and preferably at each hospital or even each unit. Based on this report, it is clear that the concept of 'know your bugs' has never been as crucial to guiding and optimising empirical treatment for bacteraemic infections in particular. This also applies to several other common hospital-acquired pathogens such as enterococci, where current comprehensive data on vancomycin resistance in private institutions are largely lacking. The true incidence of Clostridium difficile infections is also unknown. These challenges must all be urgently addressed to improve future private sector HAI pathogen and AMR surveillance.
The Group for Enteric Respiratory and Meningeal disease Surveillance in South Africa (GERMS-SA)
GERMS-South Africa is an active laboratory-based surveillance programme for bacterial and fungal pathogens of public health importance. Funded by the NHLS and Centers for Disease Control and Prevention (Atlanta, USA), it receives clinical isolates and specimens from a nationwide network of 270 public and private sector laboratories throughout the country. Laboratories submit clinical isolates according to specific case definitions, together with basic demographic data. In addition, enhanced surveillance activities take place at 16 sentinel sites servicing 25 hospitals. In these locations, dedicated surveillance officers collect additional clinical and epidemiological information on all laboratory-confirmed cases.
GERMS-SA has four main areas of interest, namely AIDS-related opportunistic infections, epidemic-prone diseases, vaccine-preventable diseases and nosocomial infections. The various reference units of the NICD monitor the number of cases of 11 specific bacterial and fungal organisms isolated by participating laboratories, and conduct additional laboratory phenotypic and genotypic characterisation studies. The pathogens of interest are Salmonella spp., Shigella spp., Vibrio spp. and Cryptococcus spp. isolated from any site; diarrhoeagenic E. coli isolated from a stool or rectal swab; Pneumocystis jirovecii isolated from a respiratory tract specimen; and S. pneumoniae, N. meningitidis and H. influenzae isolated from any normally sterile body site. As mentioned earlier in this section, a new reference unit has been established specifically for the study of AMR in nosocomial pathogens. This unit will focus initially on K. pneumoniae and S. aureus isolates from blood culture.
GERMS-SA conducts regular audits at participating laboratories to ensure the quality and completeness of isolates submitted. The stored isolates form a valuable isolate bank that can be accessed for additional special studies conducted periodically. GERMS-SA produces an annual report, as well as a quarterly surveillance bulletin and numerous publications. As a result of their surveillance activities, GERMS-SA has developed an extensive database relating to communicable diseases in South Africa, which is used to inform public health decision making.
Enteric Diseases Reference Unit
The Enteric Diseases Reference Unit (EDRU) at the NICD was started in 1997, under the guidance of a pathologist and a part-time technologist. Currently, the EDRU participates in a national, active, laboratory-based surveillance programme through its involvement with GERMS-SA.
The EDRU collects data on patients presenting throughout South Africa with both invasive and non-invasive disease caused by diarrhoea-causing bacteria, Salmonella spp. (including S. enterica serotype Typhi, hereafter referred to as S. Typhi), Shigella spp., V. cholerae and diarrhoeagenic E. coli that meet the EDRU's predetermined case definitions. The EDRU collates all the patient and isolate information in a single record and it is these data that GERMS-SA is able to report on. The EDRU under GERMS-SA have patient and isolate records captured into a secure electronic database from 2003 to the present.
In an attempt to make these data representative and reflective of the disease burden in each province in the country, all diagnostic laboratories throughout the country are motivated to voluntarily submit limited demographic details and isolates to the EDRU. In exchange, the EDRU offers serogrouping, serotyping and AST at no cost. Epsilometer tests (E-tests) are used to determine the MIC of each isolate to antimicrobial agents, according to CLSI, formerly the National Committee on Clinical Laboratory Standards (NCCLS), guidelines.
The unit has the capacity to perform genotypic characterisation of isolates, which is particularly useful in outbreak situations. The molecular epidemiology of these bacterial pathogens is continually being elucidated, specifically that of outbreak or epidemic-prone pathogens such as S. Typhi, Shigella dysenteriae type 1 and V. cholerae. A multiplex polymerase chain reaction (M-PCR) is used to identify the presence of toxin genes in diarrhoeagenic E. coli. In addition the EDRU's molecular research laboratory is involved with characterising the molecular basis for AMR in these pathogens.
STI Reference Centre
While no STI surveillance systems exist in the private sector, the numbers of total STI syndrome episodes and new episodes of male urethritis syndrome (MUS) are recorded at all public sector primary health care clinics (PHCs); however, no data are routinely recorded for other STI syndromes. For this reason, a national sentinel clinical STI syndrome surveillance system was launched in November 2003. This surveillance system, designed by the STI Reference Centre and implemented in collaboration with the NDoH, operates at 270 clinical sites across South Africa. The STI Reference Centre analysed and reported the data for the first year of the sentinel survey (April 2004 -March 2005); subsequent to this, the clinically based sentinel surveillance system has been managed in its entirety by the NDoH.
The STI Reference Centre is part of the NICD, a division of the parastatal NHLS established in 2001. The current activities of the STI Reference Centre are in keeping with the mission of the NICD, which is to be a resource of knowledge and expertise in regionally relevant communicable diseases to the South African Government, to Southern African Development Community countries and to the African continent at large, in order to assist in the planning of policies and programmes and to support appropriate responses to communicable disease issues. The STI Reference Centre's main operational focus concerns STI surveillance, research, training and teaching. The Centre's current goals are to strengthen microbiological surveillance in South Africa and to establish, in collaboration with the World Health Organization (WHO), a Gonococcal Antimicrobial Surveillance Programme (GASP) network across Africa to provide a more complete regional AMR profile for STIs.
The Centre has performed aetiological and AMR surveys in most of South Africa's nine provinces over the past 5 years. Patients with MUS, vaginal discharge syndrome (VDS) and genital ulcer syndrome (GUS) with informed written consent provide anonymous samples, labelled with a unique survey number, for laboratory work-up. All patients who are enrolled into surveys receive syndromic treatment for their STIs, are given contact slips for partner notification and are offered on-site HIV counselling and testing.
For MUS or VDS patients, urine or urethral swabs (men) or endocervical swabs (women) are collected to detect Neisseria gonorrhoeae, Chlamydia trachomatis, Trichomonas vaginalis and Mycoplasma genitalium by real-time M-PCR assay. Vaginal smears from VDS cases are Gram-stained to detect the presence of Candida spp. and/or the presence of bacterial vaginosis. Ulcer swabs are tested for herpes simplex virus (HSV), Treponema pallidum, Haemophilus ducreyi and C. trachomatis L1-L3 by real time M-PCR. Giemsa-stained ulcer smears are examined to diagnose granuloma inguinale. Syphilis, herpes simplex virus (HSV) type 2 and HIV serology is additionally performed on sera from each patient.
AST for bacterial STI pathogens is only performed with N. gonorrhoeae isolates cultured from urethral swabs. Following presumptive and confirmatory identification, MICs are determined for cefixime, ceftriaxone and ciprofloxacin by E-test. The gonococci are also stored in cryovials, preferably at -70ºC, and transferred to the STI Reference Centre at the end of each survey for subsequent agar dilution MIC determinations using a wider panel of antimicrobial agents.
The STI Reference Centre is playing a leading role in the development of GASP in Africa, which will feed into the WHO's global GASP. In relation to GASP activities, the Centre first assisted the Namibian Ministry of Health and Social Services to conduct aetiological and AMR surveillance in 2007. At present, the Centre is supporting health ministries and laboratories in Zimbabwe, Madagascar and Tanzania with ongoing or planned AMR surveys in terms of technical assistance with protocol writing and training of both laboratory and clinical staff.
Conclusion
To address the challenge of increasing resistance in these diseases, it will be necessary to begin AMR testing for a wider range of organisms, possibly following the GASP model. Because these pathogens are easily transmitted, it is particularly important that clinicians prescribe effective antibiotics capable of eradicating the pathogen during infection. This is particularly important for strains resistant to other antimicrobials. As most prescribing for these infections is empirical, an important element in appropriate prescribing is knowledge of resistance. It is therefore important that comprehensive laboratory surveillance of these diseases, sufficient to provide data representative of national disease epidemiology, is undertaken to monitor changes in AMR, particularly the evolution of MDR.
References
1. Brink AJ, Moolman J, Cruz da Silva M, and the National Antibiotic Surveillance Forum. Antimicrobial susceptibility profile of selected bacteraemic pathogens from private institutions in South Africa. S Afr Med J 2007;97:630-636. [ Links ]
2. Brink AJ, Feldman C, Richards GA, and the National Antibiotic Surveillance Forum. Emergence of extensive drug resistance (XDR) among Gram-negative bacilli in South Africa - moving a step closer. S Afr Med J 2008;98:586-592. [ Links ]














