Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.120 n.1-2 Pretoria Jan./Feb. 2024
http://dx.doi.org/10.17159/sajs.2024/13211
REVIEW ARTICLE
South African actinobacteria: A treasure trove of novel bioactive metabolites for drug discovery
Kojo S. AcquahI, II; David W. GammonI; Denzil R. BeukesIII
IDepartment of Chemistry, University of Cape Town, Cape Town, South Africa
IIDivision of Medicinal Chemistry, Department of Pharmaceutical Sciences, University of Connecticut, Storrs, Connecticut, USA
IIISchool of Pharmacy, University of the Western Cape, Cape Town, South Africa
ABSTRACT
Although South Africa is known as one of the most biodiverse countries in the world, based on its unique plants and animals, microorganisms have received much less attention. Microorganisms in general and actinobacteria in particular are an underexplored source of new medicines. Recent studies have demonstrated the presence of diverse cultivable actinobacteria from various biomes. However, investigations of the natural product diversity associated with these microorganisms are lacking. We hereby present a review of natural products isolated from South African actinobacteria together with their biological activities. Many of these natural products are structurally novel and include compounds belonging to the following classes: anthraquinones, isoflavonoids, ketolides, macrolides, macrolactams, tripeptides and depsipeptides. They show a wide range of biological activities including antibacterial, antifungal, cytotoxic and antitumour activities.
SIGNIFICANCE:
• This review highlights the importance of actinobacteria in the discovery of new medicines and summarises the state-of-the-art on their research in South Africa.
• We reveal a gap in the exploitation of this resource and emphasise the opportunities for multidisciplinary research.
Keywords: Actinomycetes, filamentous bacteria, microbial biodiversity, natural products, biological activity
Introduction
Natural products from plants, invertebrates and microorganisms have played an important role in the development of new medicines and agrochemicals.1 Microbial natural products, in particular, offer significant advantages over natural products produced by macroorganisms. These advantages include a reduced impact on the environment (ecosystems), and hence reduced competition with food crops for arable land, as well as relative ease of production and manipulation of biosynthetic pathways to produce novel compounds for commercial exploitation.
As one of the most biodiverse countries in the world2, South Africa has a rich tradition of natural products research.3 However, the main focus of these endeavours has been on plant natural products, and more recently marine natural products4,5, with much less attention on microbial natural products.
One of the most important sources of medicinally important natural products are the actinobacteria. Actinobacteria, also known as actinomycetes, are filamentous Gram-positive bacteria with high guanine and cytosine (G+C) content in their DNA. The phylum Actinobacteria comprises 20 orders and more than 50 families.6 This bacterial phylum is widely distributed in terrestrial and both fresh and marine aquatic environments. Some can thrive under extreme conditions such as hyperaridity, high salinity, cold, high pressure, low pH (acidic) and heavy metal contaminated ecosystems.7 Actinobacteria are either free living, such as soil-dwelling bacteria, or living in association with other organisms, like the plant commensals or those living in and/or on the surfaces of animals like ants, termites and marine invertebrates.8-10 Some actinobacteria are also plant and animal pathogens11, while others find use in agriculture, biotechnology, and medicine. In agriculture, they are saprophytic and break down dead plant and animal remains, hastening decomposition.8 They also aid in nitrogen fixation and are known to produce plant growth promoters, insecticides, herbicides and fungicides.9 Natural products produced by actinobacteria are structurally diverse and have shown diverse biological activities, including antioxidant, antimalarial, anthelmintic, antifungal, enzyme inhibitory, antibacterial, anticancer, immunosuppressive and cardiovascular properties.9,12,13 Antibiotics are the largest class of drugs discovered from actinomycetes, as they produce about 70% of all naturally derived antibiotics currently in clinical use.9 Most of these antibiotics were discovered during the 'golden era' of antibiotic drug discovery and include the aminoglycosides, β-lactams, glycopeptides, macrolides, rifamycins and tetracyclines.12
In this review, we discuss the natural products produced by actinobacterial strains isolated from South African environments. The many excellent studies focusing only on the distribution and description of new actinobacteria species, as well as those only reporting on the biological activity of crude extracts and the discovery of enzymes fall outside the scope of this review.
Natural products from South African actinobacteria
Several novel actinomycete strains have been isolated from South African soils, flora and fauna, in both terrestrial and marine environments, and have been shown to contain bio-active secondary metabolites (Table 1). These strains include species of the ubiquitous Streptomyces genus and the less isolated rare genera Actinomadura, Actinosynnema, Amycolatopsis, Gordonia, Kribbella, Nocardia, Nonomuraea, Rhodococcus, Streptosporangium, Saccharopolyspora and Tsukamurella.14-22
Streptomyces
The first report of a South African actinomycete-derived secondary metabolite was a tetraene macrolide natamycin 1 (Figure 1) (also known as pimaricin, natacyn, tennecetin and E235) which was patented in 1955 for its antifungal activity.23,24 This antibiotic was first purified in 1955 from the extract of the culture broth of Streptomyces natalensis, isolated from a soil sample collected from Pietermaritzburg (giving rise to its original name, pimaricin) in the KwaZulu-Natal Province, South Africa.24
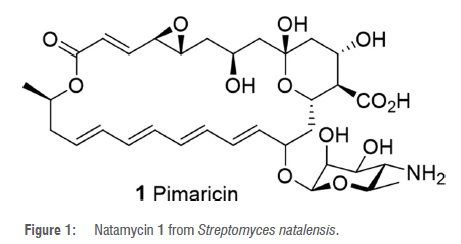
Natamycin 1 was later also isolated from Streptomyces gilvosporeus, Streptomyces chattanogenesis and Streptomyces lydicus2521It is active against a variety of saprophytic and parasitic fungi and is therefore used commercially as a preservative.28 It acts by binding to ergosterol in the fungal cell wall, thereby inhibiting fungal growth.29,30 Natamycin 1 therefore has a wide spectrum of antifungal activity and minimal toxicity to mammalian cells. It also displays in vitro activity against numerous protozoa including Trypanosoma and Acanthamoeba.3132Clinically, natamycin 1 is used to treat keratitis, and especially that caused by Aspergillus fumigatus, Candida albicans and Acanthamoeba sp.28,32 It is also used to treat fungal infections caused by Cephalosporium, Fusarium and Penicillium, and has shown activity against Alternaria, Colletotrichum, Curvularia, Lasiodiplodia, Scedosporium and Trichophyton.28It is a commercial food additive which has been used for about half a century to prevent fungal growth on foods such as cheese, sausages, yoghurt, fruits, meats, baked confectioneries and beverages.28
A soil microbe antibiotic screening programme led to the isolation of actinomycete strain AB 1246E-26 from South African bushveld soil.33 Although the genus of this actinomycete strain was not determined, preliminary characterisation narrowed the taxonomic assignment to either Nocardia or the defunct Micropolyspora.33Strain AB 1246E-26 showed activity against the antibiotic-sensitive strain of Pseudomonas aeruginosa K799/61 among other P aeruginosa strains.33,34 The organic extract of the whole fermentation broth of strain AB 1246E-26 was subjected to a bioactivity guided isolation protocol to yield the novel anthraquinone-derived class of antibiotics called altromycins.34,35 The altromycins A-I 2-10 (Figure 2) are nine closely related members of the pluramycin class of compounds with a single epoxide substituent, an amino-disaccharide and/or a 6-deoxy-3-O-methylaltrose attached to the conjugated ring systems of an anthraquinone-y-pyrone core.34-36 Altromycin B 3 was screened against 30 bacterial strains including Staphylococcus aureus, Staphylococcus epidermidis, Micrococcus luteus, Enterococcus hirae, Streptococcus bovis, Streptococcus agalactiae, Streptococcus pyogenes, Escherichia coli, Enterobacter aerogenes, Klebsiella pneumoniae, Providencia stuartii, Pseudomonas aeruginosa, Pseudomonas cepacia and Acinetobacter sp. clinical isolates.33 Altromycin B 3 exhibited potent antibacterial activity against the clinical isolates of Staphylococcus and Streptococcus with a minimum inhibitory concentration (MIC) range of 0.39-3.12 μg/mL and 0.2-3.12 μg/mL, respectively, but displayed moderate to weak antibacterial activity against Gram-negative bacteria with an MIC range of 25 to >100 μg/mL.33 The altromycins also showed cytotoxic activity against various cancer and tumour cell lines including cervical cancer (HeLa), human lung cancer (A549), colon tumour (HCT-8), murine leukemia cell (P388) lines and ovarian sarcoma (M5076).37
In their search for antibiotics that inhibit fatty acid biosynthesis in bacteria, researchers at Merck discovered the novel broad-spectrum antibiotic platensimycin 11 (Figure 3) from Streptomyces platensis strain MA7327, which was originally isolated from a soil sample collected in the Eastern Cape Province of South Africa.38 An organic extract of the fermentation broth of S. platensis strain MA7327 was subjected to a unique antisense differential sensitivity whole-cell two-plate agar diffusion bioassay-guided fractionation process to yield platensimycin 11.38Platensimycin 11 consists of a 3-amino-2,4-dihydroxybenzoic acid tethered via an amide bond to a C-17 tetracyclic enone which includes a bridge-head oxygen.38,39 Another closely related compound, platencin 12 (Figure 3), was produced by Streptomyces platensis strain MA7339 using the same bioassay-guided fractionation procedure, although its biosynthetic gene cluster was also identified in strain MA7327.40 Several other analogues 13-49 (Figure 3) - with modifications on or loss of the aromatic ring, modifications on the terpenoid and anilide moieties and a change in the length of the enone acid portion of platensimycin and platencin - have also been isolated from strain MA7327.41-48 Compounds 50 and 51 (Figure 3), which are structurally different from the platensimycin group of compounds, were also isolated from strain MA7327.41,42 Furthermore, other glycosylated analogues of platensimycin 11 and platencin 12 have been produced by an engineered mutant strain S. platensis SB12600.40 Platensimycin 11 selectively inhibits the elongation-condensing enzyme FabF of the bacterial fatty acid synthesis pathway, while platencin 12 equally inhibits both the initiation condensing (FabH) and elongation (FabF) enzymes.39 Platensimycin, platencin and their analogues have shown potent in vitro activity against both cell-free and whole-cell systems including methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus and Mycobacterium tuberculosis.39
Although the isolated analogues and synthesised ones did not exhibit improved activity compared with compounds 11 and 12, they provide important structure-activity relationship information to determine the pharmacophore of 11 and 12.
In their quest to isolate biologically active metabolites from termite-associated actinobacteria, Kim et al. discovered that Streptomyces strain M56, isolated from the fungal comb of a South African Macrotermes natalensis Mn802 colony, exhibited potent broad-spectrum antifungal activity.49 Bioassay-guided isolation resulted in the purification of the novel fused bicyclic ansa macrolide natalamycin A 52 (Figure 4) alongside other ansa macrolides including reblastatin 53, geldanamycin 54 and its derivates, 17-O-demethyl-geldanamycin 55, 19-S-methylgeldanamycin 56, 17-amino-17-demethoxy-geldanamycin 57, methyl geldanamycinate derivative 58, 17-amino-17-demethoxy-methyl geldanamycinate 59 and 19-[(1 'S,4'R)-4'-hydroxy-1 '-methoxy-2'-oxopentyl]geldanamycin 60.49 Although the fractions that yielded the isolated compounds showed strong antifungal activity against some strains, the natalamycins exhibited weak or no activity against Saccharomyces cerevisiae and other fungal isolates.49 An untargeted dereplication of the liquid chromatography-mass spectrometry (LCMS) data of the methanol extract of Streptomyces strain M56 signified the presence of new metabolites.50 The methanol extract of a large-scale culture of strain M56 was subjected to several chromatographic methods to yield the fused 5-8-5 tricyclic diterpene 17-hydroxycyclooctatin 61 (Figure 4).50 Compound 61 is a potential ERa antagonist and exhibited weak cytotoxicity activity against MCF-7 human breast cancer cell lines with an IC50 value of 566.95 ± 0.48 μΜ.50
Further studies on the metabolites of the actinobacteria associated with the fungus-growing South African termite M. natalensis led to the isolation of Streptomyces strain RB1 which exhibited antibacterial activity against Staphylococcus aureus and Candida albicans.51Fractionation of the methanol (MeOH) extract of strain RB1 yielded the new isoflavonoid glycosides, termisoflavone A-C 62-64. and other isoflavonoids 66-70, 72-74 (Figure 5).51 The isolated compounds showed no antifungal or antibacterial activity when screened against C. albicans, C. neoformans, S. aureus, and E. coli, but compounds 69 and 73 ameliorated cisplatin-induced kidney cell damage.51 Further investigation of the MeOH extract of strain RB1 using LCMS- and NMR-based dereplication strategies led to the identification and subsequent isolation of another new isoflavonoid glycoside, termisoflavone D 65, together with the known isoflavonoids 66, 67, 69, 71-73 and 75 (Figure 5).52 Isoflavonoid 69 displayed activity against glutamate-induced HT22 cells by preventing accumulation of intracellular reactive oxygen species.52 Another study exploring the termite associated actinobacteria for reno- and kidney-protective drug discovery found that the MeOH extract of Streptomyces sp. RB1 exhibited a protective effect against cisplatin-induced cytotoxicity.53 A bioassay (LLC-PK1 cells)-guided isolation process yielded the renoprotective 1-O-(2-aminobenzoyl)-a-L-rhamnopyranoside (ABR) 76 (Figure 5).53
Analysing the chemical and metabolomic profiles of actinobacteria derived from termite nests with an unbiased high-throughput highperformance liquid chromatography-high-resolution mass spectrometry based dereplication strategy revealed that Streptomyces sp. M41 isolated from the South African termite M. natalensis produces new complex nonribosomal peptide polyketide synthase (NRPS/PKS) hybrid compounds.54 Chromatographic purification of a large-scale culture of strain M41 interestingly yielded new analogues (two linear 77, 78 and one cyclic 79) of the cyclic depsipeptide dentigerumycin 80 (Figure 6).54
The South African soil actinomycete, Streptomyces canus strain CA-091830, is a producer of the cyclic depsipeptides krisynomycins A-C 81-83 (Figure 7).55,56 Krisynomycin A was initially isolated based on a screening project with the aim of identifying and isolating imipenem potentiators against methicillin-resistant Staphylococcus aureus (mRsA).55 Further investigation led to the isolation of Krisynomycin B and C, which are chlorinated analogues of Krisynomycin A.56 Although compounds 81-83 showed weak activity against MRSA, the activity was improved when they were tested in combination with sub-lethal concentrations of imipenem.56
Rare actinomycete strains
The rare actinomycete Actinomadura sp. 5-2, which was recovered from the gut of the fungus-growing termite M. natalensis, produced novel, highly substituted tropolone alkaloids, rubterolones A-F 84-89 (Figure 8).57 These compounds were detected by both bioactivity and high-resolution mass spectrometry based dereplication techniques, and subsequently isolated from the organic extract of a culture of strain 5-2.57 Curiously, compounds 84-89 did not show any significant antifungal activity.57
The crude extract of another Actinomadura isolate, strain RB99, isolated from the surface of the termite M. natalensis, was analysed by liquid chromatography (LC)/ultraviolet (UV)/mass spectrometry (MS) and shown to produce new compounds.58 A spectrometry guided isolation led to the discovery of three new cyclic tripeptides named natalenamides A-C 90-92 (Figure 9).58 The isolated compounds exhibited weak cytotoxicity when screened against HepG2 and HeLa/A549 cells.58 Compound 92 showed significant activity against IBMX-mediated melanin synthesis in a dose-dependent manner.58
Analyses of the high-resolution tandem mass spectrometry (HR-MS2) data of the MeOH extract of strain RB99 and further exploration of the HR-MS2 data on the Global Natural Product Social (GNPS) molecular networking platform showed that strain RB99 produces polyhalogenated isoflavonoids.59 Seoung et al. proved that Actinomadura sp. RB99 can bio-transform the plant-based daidzein 74 and genistein 75 isoflavonoids contained in the ISP2 growth medium to polyhalogenated derivatives.59 Optimisation of the growth medium (ISP2 augmented with NaCl or KBr) led to the production and subsequent MS-guided purification of eight polychlorinated analogues 93-100 (Figure 9), of which six were new, and seven novel polybrominated analogues 101-107 (Figure 9), of daidzein and genistein.59 The isolated chlorinated analogues did not exhibit any antibacterial or antifungal activities against E. coli, S. aureus, S. epidermidis and C. albicans, but the brominated analogues 101 and 105 were active against Helicobacter pylori.59Additional analysis of the LCMS data of the MeOH extract of strain RB99 led to the detection and isolation of the antibiotic and antitumour agent fridamycin A 108 (Figure 9), which is a type II polyketide.60 Fridamycin A 108 showed good antidiabetic properties in 3T3-L1 adipocytes and could serve as a promising lead for type 2 diabetes drug discovery.60
Metabolomic and bioactivity profiling of termite-associated actinomycetes led to the detection and subsequent isolation of four new 20-membered glycosylated polyketide macrolactams, named macrotermycins A-D 109-112 (Figure 10), from the organic extract of the rare actinomycete Amycolatopsis sp. M39.61 Strain M39 was also isolated from the termite M. natalensis and its organic crude extract exhibited a unique metabolomic profile and was active against the termite fungal garden competitor Pseudoxylaria spp.61 Only compounds 109 and 111 were active against Pseudoxylaria sp.61
A rare actinomycete, Kribbella speibonae strain SK5, isolated from a soil sample collected from Stellenbosch in the Western Cape Province of South Africa, displayed strong antimycobacterial activity against Mycobacterium aurum strain A+.62 Chemical and metabolomic profiling of an organic extract of a liquid culture of this strain showed that it is a prolific producer of hydroxamate siderophores, including new dehydroxylated desferrioxamine analogues and diketopiperazines (DKP).63 Two new dehydroxylated desferrioxamines, speibonoxamine 113 and desoxy-desferrioxamine D1 114 (Figure 11), alongside already reported desferrioxamines 115-118 and a DKP 119 (Figure 11), were subsequently isolated from an organic extract of a liquid culture of strain SK5.63 The plausible structures of three new dehydroxylated analogues 120-122 were determined by the GNPS molecular network and MS/MS fragmentation analyses.63
Conclusions and future prospects
Biodiversity is "more than just legs and leaves"64 and South Africa's microbial biodiversity presents a tremendous opportunity for the natural products chemist and those interested in drug discovery. In this review, we have described 122 compounds and shown that South African actinobacteria are prolific producers of novel, bioactive metabolites. Interestingly, the first compound described from a South African actinomycete, natamycin, is also the only one that has made it to the clinic. Other compounds, such as platensimycin and geldanamycin, have shown promise but either lack efficacy in humans or showed toxic side effects which prevented their development as drugs. South African researchers interested in natural products based drug discovery face the same challenges as elsewhere in the world.
These challenges include the significant cost of drug development, re-isolation of previously reported compounds, and lack of interest in natural products for drug development. Nevertheless, the compounds reviewed here present only the tip of the iceberg and many more species remain to be discovered and studied for natural product production. Furthermore, with innovations and technological advancement in purification, structure elucidation, chemical biology, genome sequencing and mining, dereplication, and bioinformatic, cheminformatic and metabolomic tools like the GNPS molecular networking, microbial natural product drug discovery in South Africa shows great potential. It is worth mentioning that, apart from our research on the chemistry of the metabolites of the South African rare actinomycete Kribbella speibonae strain SK5, all the research on South African actinomycetes reported here was done by research groups based outside of South Africa. This represents a challenge and an opportunity for closer collaboration between South African researchers (microbiologists, pharmacologists and chemists) in order to fully explore the opportunities presented by South African microbial biodiversity.
Acknowledgements
We gratefully acknowledge financial support from the National Research Foundation of South Africa (grant no. 138000).
Competing interests
We have no competing interests to declare.
Authors' contributions
K.S.A: Conceptualisation; data collection; writing - the initial draft.
D.W.G: Conceptualisation; student supervision; writing - revisions.
D.R.B: Conceptualisation; data collection; student supervision; writing -revisions.
References
1. Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770-803. https://doi.org/10.1021/acs.jnatprod.9b01285 [ Links ]
2. UN Environment Programme World Conservation Monitoring Centre (UNEP-WCMC). Biodiversity A-Z. Cambridge, UK: UNEP-WCMC; 2014. Available from: https://www.biodiversitya-z.org/content/megadiverse-countries [ Links ]
3. Drewes SE. Natural products research in South Africa: 1890-2010. S Afr J Sci. 2012;108(5/6), Art. #765. https://doi.org/10.4102/sajs.v108i5/6.765 [ Links ]
4. Davies-Coleman MT. Natural products research in South Africa: End of an era on land or the beginning of an endless opportunity in the sea? S Afr J Chem. 2010;63:105-113. [ Links ]
5. Davies-Coleman MT, Veale CG. Recent advances in drug discovery from South African marine invertebrates. Mar Drugs. 2015;13(10):6366-6383. https://doi.org/10.3390/md13106366 [ Links ]
6. Parte AC, Sardá Carbasse J, Meier-Kolthoff JP Reimer LC, Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70:5607-5612. https://doi.org/10-1099/ijsem.0.004332 [ Links ]
7. Merino N, Aronson HS, Bojanova DPP Feyhl-Buska J, Wong ML, Zhang S, et al. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front Microbiol. 2019;10, Art. #780. https://doi.org/10.3389/fmicb.2019.00780 [ Links ]
8. Mayfield CI, Williams ST, Ruddick SM, Hatfield HL. Studies on the ecology of actinomycetes in soil IV. Observations on the form and growth of Streptomycetes in soil. Soil Biol Biochem. 1972;4(1):79-91. https://doi.org/10.1016/0038-0717(72)90045-4 [ Links ]
9. Barka EA, Vatsa P Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):143. https://doi.org/10.1128/MMBR.00019-15 [ Links ]
10. Benndorf R, Martin K, Küfner M, de Beer ZW, Vollmers J, Kaster AK, et al. Actinomadura rubteroloni sp. nov. and Actinomadura macrotermitis sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis. Int J Syst Evol. 2020;70(10):5255-5262. https://doi.org/10.1099/ijsem.0.004403 [ Links ]
11. Salwan R, Sharma V. The role of actinobacteria in the production of industrial enzymes. In: Singh BP, Gupta VK, Passari AK, editors. New and future developments in microbial biotechnology and Bioengineering. Amsterdam: Elsevier; 2018. p. 165-177. https://doi.org/10.1016/B978-0-444-63994-3.00011-4 [ Links ]
12. Genilloud O. Actinomycetes: Still a source of novel antibiotics. Nat Prod Rep. 2017;34(10):1203-1232. https://doi.org/10.1039/C7NP00026J [ Links ]
13. Taechowisan T, Chaisaeng S, Phutdhawong WS. Antibacterial, antioxidant and anticancer activities of biphenyls from Streptomyces sp. BO-07: An endophyte in Boesenbergia rotunda (L.) Mansf A. Food Agric Immunol. 2017;28(6):1330-1346. https://doi.org/10.1080/09540105.2017.1339669 [ Links ]
14. Rohland J, Meyers PR. Streptomyces fractus sp. nov., a novel streptomycete isolated from the gut of a South African termite. Antonie van Leeuwenhoek. 2015;107(5):1127-1134. https://doi.org/10.1007/s10482-015-0404-8 [ Links ]
15. Cook AE, Le Roes M, Meyers PR. Actinomadura napierensis sp. nov., isolated from soil in South Africa. Int J Syst Evol. 2005;55(2):703-706. https://doi.org/10.1099/ijs.0.63359-0 [ Links ]
16. Everest GJ, Le Roes-Hill M, Rohland J, Enslin S, Meyers PR. Amycolatopsis roodepoortensis sp. nov. and Amycolatopsis speibonae sp. nov.: Antibiotic-producing actinobacteria isolated from South African soils. J Antibiot. 2014; 67(12):813-818. https://doi.org/10.1038/ja.2014.79 [ Links ]
17. Everest GJ, Meyers PR. Kribbellahippodromisp. nov., isolated from soil from a racecourse in South Africa. Int J Syst Evol Microbiol. 2008;58(2):443-446. https://doi.org/10.1099/ijs.0.65278-0 [ Links ]
18. Le Roes M, Goodwin CM, Meyers PR. Gordonia lacunae sp. nov., isolated from an estuary. Syst Appl Microbiol. 2008;31(1):17-23. https://doi.org/10.1016/j.syapm.2007.10.001 [ Links ]
19. Benndorf R, Schwitalla JW, Martin K, De Beer ZW, Vollmers J, Kaster AK, et al. Nocardiamacrotermitis sp. nov. and Nocardia aurantia sp. nov., isolated from the gut of the fungus-growing termite Macrotermes natalensis. Int J Syst Evol Microbiol. 2020;70(10):5226-5234. https://doi.org/10.1099/ijsem.0.004398 [ Links ]
20. Le Roes M, Meyers PR. Nonomuraea candida sp. nov., a new species from South African soil. Antonie van Leeuwenhoek. 2008;93(1):133-139. https://doi.org/10.1007/s10482-007-9187-x [ Links ]
21. Sibanda T, Mabinya LV, Mazomba N, Akinpelu DA, Bernard K, Olaniran AO, et al. Antibiotic producing potentials of three freshwater actinomycetes isolated from the Eastern Cape Province of South Africa. Int J Mol Sci. 2010;11(7):2612-2623. https://doi.org/10.3390/ijms11072612 [ Links ]
22. Le Roes-Hill M, Durrell K, Prins A, Meyers PR. Streptosporangium minutum sp. nov., isolated from garden soil exposed to microwave radiation. J Antibiot. 2018;71(6):564-574. https://doi.org/10.1038/s41429-018-0036-0 [ Links ]
23. Struyk AP, Waisvisz JM. (Gist-Brocades, Delft): Pimaricin and process of producing same. US-Patent. 1955;3:892. [ Links ]
24. Struyk AP Drost G, Haisvisz JM, Van Eek T, Hoogerheide JC. Pimaricin, a new antifungal antibiotic. Antibiot Annu. 1957/1958;5:878-885. [ Links ]
25. Chen GQ, Lu FP Du LX. Natamycin production by Streptomyces gilvosporeus based on statistical optimization. J Agric Food Chem. 2008;56(13):5057-5061. https://doi.org/10.1021/jf800479u [ Links ]
26. Du YL, Chen SF, Cheng LX Shen XL, Tian Y Li YQ. Identification of a novel Streptomyces chattanoogensis L10 and enhancing its natamycin production by overexpressing positive regulator ScnRII. J Microbiol. 2009;47(4):506-513. https://doi.org/10.1007/s12275-009-0014-0 [ Links ]
27. Lu CG, Liu WC, Qiu JY, Wang HM, Liu T, Liu DW. Identification of an antifungal metabolite produced by a potential biocontrol actinomyces strain A01. Braz J Microbiol. 2008;39(4):701-707. https://doi.org/10.1590/S1517-83822008000400020 [ Links ]
28. Aparicio JF, Barreales EG, Payero TD, Vicente CM, De Pedro A, Santos-Aberturas J. Biotechnological production and application of the antibiotic pimaricin: Biosynthesis and its regulation. Appl Microbiol Biotechnol. 2016;100(1):61-78. https://doi.org/10.1007/s00253-015-7077-0 [ Links ]
29. Te Welscher YM, Jones L, Van Leeuwen MR, Dijksterhuis J, De Kruijff B, Eitzen G, et al. Natamycin inhibits vacuole fusion at the priming phase via a specific interaction with ergosterol. Antimicrob Agents Chemother. 2010;54(6):2618-2625. https://doi.org/10.1128/AAC.01794-09 [ Links ]
30. Te Welscher YM, Hendrik H, Balagué MM, Souza CM, Riezman H, De Kruijff B, et al. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J Biol Chem. 2008;283(10):6393-6401. https://doi.org/10.1074/jbc.M707821200 [ Links ]
31. Rolón M, Seco EM, Vega C, Nogal JJ, Escario JA, Gómez-Barrio A, et al. Selective activity of polyene macrolides produced by genetically modified Streptomyces on Trypanosoma cruzi. Int J Antimicrob Agents. 2006;28(2):104-109. https://doi.org/10.1016/j.ijantimicag.2006.02.025 [ Links ]
32. Sunada A, Kimura K, Nishi I, Toyokawa M, Ueda A, Sakata T, et al. In vitro evaluations of topical agents to treat Acanthamoeba keratitis. Ophthalmology. 2014;121(10):2059-2065. https://doi.org/10.1016/j.ophtha.2014.04.013 [ Links ]
33. Jackson M, Karwowski JP, Theriault RJ, Hardy DJ, Swanson SJ, Barlow GJ, et al. Altromycins, novel pluramycin-like antibiotics I. Taxonomy of the producing organism, fermentation and antibacterial activity. J Antibiot. 1990;43(3):223-228. https://doi.org/10.7164/antibiotics.43.223 [ Links ]
34. Brill GM, Mcalpine JB, Whittern DN, Buko AM. Altromycins, novel pluramycin-like antibiotics II. Isolation and elucidation of structure. J Antibiot. 1990;43(3):229-237. https://doi.org/10.7164/antibiotics.43.229 [ Links ]
35. Brill GM, Jackson M, Whittern DN, Buko AM, Hill P Chen RH, et al. Altromycins E, F, G, H and I; additional novel components of the altromycin complex. J Antibiot. 1994;47(10):1160-1164. https://doi.org/10.7164/antibiotics.47.1160 [ Links ]
36. Hansen M, Hurley L. Altromycin B threads the DNA helix interacting with both the major and the minor grooves to position itself for site-directed alkylation and guanine N7. J Am Chem Soc. 1995;117(9):2421-2429. https://doi.org/10.1021/ja00114a006 [ Links ]
37. McAlpine JB, Karwowski JP, Jackson M, Brill GM, Kadam S, Shen L, et al. Altromycins: A new family of antitumor antibiotics-discovery and biological evaluation. In: Valeriote FA, Corbett TH, Baker LH, editors. Anticancer drug discovery and development: Natural products and new molecular models. Boston, MA: Springer; 1994. p. 95-117. https://doi.org/10.1007/978-1-4615-2610-0_6 [ Links ]
38. Singh SB, Jayasuriya H, Ondeyka JG, Herath KB, Zhang C, Zink DL, et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J Am Chem Soc. 2006;128(36):11916-11920. https://doi.org/10.1021/ja062232p [ Links ]
39. Martens E, Demain AL. Platensimycin and platencin: Promising antibiotics for future application in human medicine. J Antibiot. 2011;64(11):705-710. https://doi.org/10.1038/ja.2011.80 [ Links ]
40. Yu Z, Rateb ME, Smanski MJ, Peterson RM, Shen B. Isolation and structural elucidation of glucoside congeners of platencin from Streptomyces platensis SB12600. J Antibiot. 2013;66(5):291-294. https://doi.org/10.1038/ja.2013.1 [ Links ]
41. Zhang C, Ondeyka J, Herath K, Jayasuriya H, Guan Z, Zink DL, et al. Platensimycin and platencin congeners from Streptomyces platensis. J Nat Prod. 2011;74(3):329-340. https://doi.org/10.1021/np100635f [ Links ]
42. Zhang C, Ondeyka J, Dietrich L, Gailliot FP Hesse M, Lester M, et al. Isolation, structure and biological activities of platencin A2-A4 from Streptomyces platensis. Bioorg Med Chem. 2010;18(7):2602-2610. https://doi.org/10-1016/j.bmc.2010.02.030 [ Links ]
43. Herath KB, Zhang C, Jayasuriya H, Ondeyka JG, Zink DL, Burgess B, et al. Structure and semisynthesis of platensimide A, produced by Streptomyces platensis. Org Lett. 2008;10(9):1699-1702. https://doi.org/10.1021/ol900695v [ Links ]
44. Singh SB, Jayasuriya H, Herath KB, Zhang C, Ondeyka JG, Zink DL, et al. Isolation, enzyme-bound structure, and activity of platensimycin A1 from Streptomyces platensis. Tetrahedron Lett. 2009;50(37):5182-5185. https://doi.org/10.1016/j.tetlet.2009.06.118 [ Links ]
45. Zhang C, Ondeyka J, Zink DL, Burgess B, Wang J, Singh SB. Isolation, structure and fatty acid synthesis inhibitory activities of platensimycin B1-B3 from Streptomyces platensis. Chem Comm. 2008(40):5034-5036. https://doi.org/10.1039/b810113b [ Links ]
46. Zhang C, Ondeyka J, Guan Z, Dietrich L, Burgess B, Wang J, et al. Isolation, structure and biological activities of platensimycin B4 from Streptomyces platensis. J Antibiot. 2009;62(12):699-702. https://doi.org/10.1038/ja.2009.106 [ Links ]
47. Singh SB, Ondeyka JG, Herath KB, Zhang C, Jayasuriya H, Zink DL, et al. Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis. Bioorg Med Chem Lett. 2009;19(16):4756-4759. https://doi.org/10.1016/j.bmcl.2009.06.061 [ Links ]
48. Jayasuriya H, Herath KB, Ondeyka JG, Zink DL, Burgess B, Wang J, et al. Structure of homoplatensimide A: A potential key biosynthetic intermediate of platensimycin isolated from Streptomyces platensis. Tetrahedron Lett. 2008;49(22):3648-3651. https://doi.org/10.1016/j.tetlet.2008.03.155 [ Links ]
49. Kim KH, Ramadhar TR, Beemelmanns C, Cao S, Poulsen M, Currie CR, et al. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem Sci. 2014;5(11):4333-4338. https://doi.org/10.1039/C4SC01136H [ Links ]
50. Lee SR, Lee D, Park M, Lee JC, Park HJ, Kang KS, et al. Absolute configuration and corrected NMR assignment of 17-hydroxycyclooctatin, a fused 5-8-5 tricyclic diterpene. J Nat Prod. 2020;83(2):354-361. https://doi.org/10.1021/acs.jnatprod.9b00837 [ Links ]
51. Kang HR, Lee D, Benndorf R, Jung WH, Beemelmanns C, Kang KS, et al. Termisoflavones A-C, isoflavonoid glycosides from termite-associated Streptomyces sp. RB1. J Nat Prod. 2016;79(12):3072-3078. https://doi.org/10.1021/acs.jnatprod.6b00738 [ Links ]
52. Lee SR, Song JH, Song JH, Ko HJ, Baek JY Trinh TA, et al. Chemical identification of isoflavonoids from a termite-associated Streptomyces sp. RB1 and their neuroprotective effects in murine hippocampal HT22 cell line. Int J Mol Sci. 2018;19(9), Art. #2640. https://doi.org/10.3390/ijms19092640 [ Links ]
53. Lee D, Kang KS, Lee HJ, Kim KH. Chemical characterization of a renoprotective metabolite from termite-associated Streptomyces sp. RB1 against cisplatin-induced cytotoxicity. Int J Mol Sci. 2018;19(1), Art. #174. https://doi.org/10.3390/ijms19010174 [ Links ]
54. Wyche TP Ruzzini AC, Beemelmanns C, Kim KH, Klassen JL, Cao S, et al. Linear peptides are the major products of a biosynthetic pathway that encodes for cyclic depsipeptides. Org Lett. 2017;19(7):1772-1775. https://doi.org/10.1021/acs.orglett.7b00545 [ Links ]
55. Therien AG, Huber JL, Wilson KE, Beaulieu P Caron A, Claveau D, et al. Broadening the spectrum of β-lactam antibiotics through inhibition of signal peptidase type I. Antimicrob Agents Chemother. 2012;56(9):4662-4670. https://doi.org/10.1128/AAC.00726-12 [ Links ]
56. Perez-Bonilla M, Oves-Costales D, González I, De la Cruz M, Martin J, Vicente F, et al. Krisynomycins, imipenem potentiators against methicillin-resistant Staphylococcus aureus, produced by Streptomyces canus. J Nat Prod. 2020;83(9):2597-2606. https://doi.org/10.1021/acs.jnatprod.0c00294 [ Links ]
57. Guo H, Benndorf R, Leichnitz D, Klassen JL, Vollmers J, Görls H, et al. Isolation, biosynthesis and chemical modifications of rubterolones A-F: Rare tropolone alkaloids from Actinomadura sp. 5-2. Chem Eur J 2017;23:9338-9345. https://doi.org/10.1002/chem.201701005 [ Links ]
58. Lee SR, Lee D, Yu JS, Benndorf R, Lee S, Lee DS, et al. Natalenamides A-C, cyclic tripeptides from the termite-associated Actinomadura sp. RB99. Molecules. 2018;23(11), Art. #3003. https://doi.org/10.3390/molecules23113003 [ Links ]
59. Rak Lee S, Schalk F, Schwitalla JW, Benndorf R, Vollmers J, Kaster AK, et al. Polyhalogenation of isoflavonoids by the termite-associated Actinomadura sp. RB99. J Nat Prod. 2020;83(10):3102-3110. https://doi.org/10.1021/acs.jnatprod.0c00676 [ Links ]
60. Yoon SY, Lee SR, Hwang JY, Benndorf R, Beemelmanns C, Chung SJ, et al. Fridamycin A, a microbial natural product, stimulates glucose uptake without inducing adipogenesis. Nutrients. 2019;11(4), Art. #765. https://doi.org/10.3390/nu11040765 [ Links ]
61. Beemelmanns C, Ramadhar TR, Kim KH, Klassen JL, Cao S, Wyche TP, et al. Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org Lett. 2017;19(5):1000-1003. https://doi.org/10.1021/acs.orglett.6b03831 [ Links ]
62. Curtis SM, Norton I, Everest GJ, Pelser JG, De Kock MC, Meyers PR. Development of a Kribbella-specific isolation medium and description of Kribbella capetownensis sp. nov. and Kribbella speibonae sp. nov., isolated from soil. Antonie van Leeuwenhoek. 2020;113(5):617-628. https://doi.org/10.1007/s10482-019-01365-6 [ Links ]
63. Acquah KS, Beukes DR, Warner DF, Meyers PR, Sunassee SN, Maglangit F, et al. Novel South African rare actinomycete Kribbella speibonae strain SK5: A prolific producer of hydroxamate siderophores including new dehydroxylated congeners. Molecules. 2020;25(13), Art. #2979. https://doi.org/10.3390/molecules25132979 [ Links ]
64. Cowan DA, Valverde A, Wingfield MJ, Rybicki EP Tuffin MI. Biodiversity: So much more than legs and leaves. S Afr J Sci. 2013;109(11/12), Art. #a0037. https://doi.org/10.1590/sajs.2013/a0037 [ Links ]
 Correspondence:
Correspondence:
Denzil Beukes
Email: dbeukes@uwc.ac.za
Received: 06 Feb. 2022
Revised: 30 Aug. 2023
Accepted: 11 Sep. 2023
Published: 30 Jan. 2024
Editors: Priscilla Baker Amanda-Lee Manicum
Funding: South African National Research Foundation (grant no. 138000)














