Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.119 no.11-12 Pretoria Nov./Dez. 2023
http://dx.doi.org/10.17159/sajs.2023/15115
RESEARCH ARTICLE
https://doi.org/10.17159/sajs.2023/15115
SNP-based marker-assisted selection for high provitamin A content in African cassava genetic background
Esperance D. CodjiaI, II; Bunmi OlasanmiI, III; Chike E. UgojiII; Ismail Y RabbiII
IPan African University for Life and Earth Sciences Institute (including Health and Agriculture), University of Ibadan, Ibadan, Nigeria
IIInternational Institute of Tropical Agriculture (IITA), Ibadan, Nigeria
IIIDepartment of Crop and Horticultural Sciences, University of Ibadan, Ibadan, Nigeria
ABSTRACT
Vitamin A deficiency (VAD) contributes to significant levels of mortality and morbidity, particularly among children and women in Africa. Cassava is a major staple crop whose biofortification with beta-carotene can contribute to reducing the VAD prevalence in a cost-effective and sustainable approach. Developing high provitamin A content (pVAC) cassava varieties through the conventional approach is a laborious and slow process, partly due to the breeding bottlenecks caused by the biology of the crop. To complement the phenotypic screening for pVAC and increase selection efficiency as well as accuracy, we employed four Kompetitive Allele-Specific PCR (KASP) assays to predict the level of carotenoids in a cassava population developed from open-pollinated crosses. There was significant correlation (r = 0.88) between total carotenoid content (TCC) and root tissue colour score in the study population. Marker S1 24155522 at the phytoene synthase gene explained most of the phenotypic variation in TCC and root colour (R2 = 0.37 and 0.55, respectively) among the genotypes evaluated in this study. The other markers did not individually account for much phenotypic variation in the trait in our study population. Three genotypes - namely UIC-17-679, UIC-17-1713, and UIC-17-2823 - had higher TCCs, ranging from 10.07 μg/g to 10.88 μg/g, than the national yellow check variety IITA-IBA-TMS070593 (9.20 μg/g). Marker PSY572/S124155522 is therefore recommended for routine use in marker-assisted selection for pVAC enhancement in African cassava germplasm.
SIGNIFICANCE:
We evaluated the performance of the SNP markers associated with provitamin A content in a cassava population and draw relevant conclusions that will foster the applications of these markers in different cassava improvement programmes with similar interests. Marker-assisted selection was sufficiently accurate for an early screening of individuals for carotenoid content, especially when thousands of genotypes are usually handled. This screening will reduce efficiently the challenges and burden attached to the use of sophisticated instruments for carotenoid quantification (e.g. HPLC and I-check) for the benefit of breeders and researchers in the field.
Keywords: vitamin A deficiency, marker-assisted selection, provitamin A content, Kompetitive Allele-Specific PCR
Introduction
Inadequate micronutrient intake is a major health challenge, particularly in developing countries.1,2 This problem is exacerbated by a restricted diet that relies mostly on a single major staple crop for subsistence3,4 or on a few crops lacking some essential nutrients. About 56% of reported childhood deaths in developing nations are due to nutritional deficiencies.1 Vitamin A deficiency (VAD) causes night blindness and increases the risk of sickness and mortality in children and pregnant women.5 Globally, more than 250 000 children become blind every year, of whom half die within 12 months of losing their sight.5 In Nigeria, about 83% of children under five are vitamin A deficient.6 Supplement-based strategies for treating vitamin A deficiencies have had limited success, in part due to the short-term nature of their interventions and the inability to purchase fortified foods.4 A cost-effective approach to reducing the alarming rate of vitamin A deficiency in developing countries is through biofortification of staple crops.7
Cassava (Manihot esculenta Crantz) is an important staple crop that provides for the calorie needs of more than 800 million people in Africa, Asia and South America.8,9 Unlike other staple crops, cassava's starchy roots are inexpensive, which encourages its widespread consumption and adoption, especially in Africa.10,11 However, there is a reported correlation between consumption levels of white cassava storage roots and micronutrient deficiency, putting populations that rely on this crop at risk of concealed hunger.12 Consequently, biofortification of cassava with provitamin A content (pVAC) would contribute, non-exclusively, to reducing the VAD prevalence in West Africa4,13, and also play a crucial role in mitigating the rate of hidden hunger - a concept which refers to micronutrient deficiency with invisible symptoms in affected individuals14,15.
Significant efforts have been undertaken to exploit the natural variability for carotenoid content in the existing cassava gene pool.16-18 For instance, HarvestPlus programme and its partners have improved and released high vitamin A cassava in Nigeria in the past years. Between 2011 and 2014, two waves of yellow cassava varieties were released in Nigeria, with carotenoid contents ranging from 6 to 8 μg/g and 8 to 10 μg/g, respectively.19 The existing pVAC cassava varieties can supply up to 40% of the recommended daily intake of vitamin A.20 Nonetheless, there is a need to breed for higher pVAC cassava varieties as the breeding target fixed at 15 μg/g in yellow cassava genotypes has not yet been achieved in West Africa.17,21
The use of molecular markers to complement the conventional breeding approach is expected to increase and accelerate the genetic gain across environments and seasons22,23 compared with conventional breeding which depends on the environment24 and relies upon identification of the traits at maturity25. Moreover, known phenotyping protocols used for carotenoid quantification, such as high-performance liquid chromatography (HPLC) and I-check, are sophisticated, time-consuming and expensive21, thereby limiting the number of samples that can be handled within a short period without introducing errors at the early stage of selection when thousands of genotypes usually are handled.
Advances in molecular biology technology led to the discovery of single nucleotide polymorphism (SNP) markers which are prioritised in crop breeding due to their density in the whole genome and their need in advanced genotyping platforms.26 The commonly used SNP genotyping platforms include GoldenGate, BeadXpress and Kompetitive Allele Specific PCR, the genotyping platform used in this study. Kompetitive Allele Specific PCR (KASP) is a simple and cost-effective genotyping system, convenient for studies that involve relatively few markers.27
Efficient biofortification of cassava storage roots using the available natural variation requires a prior understanding of the genetic architecture of the trait28 and identification of quantitative trait loci (QTL)-linked markers for marker-assisted selection (MAS) implementation. A biparental QTL mapping using genotyping by sequencing was first conducted by Rabbi et al.29 and enabled the identification of a major locus on chromosome 1 which explained 92.85% of the variation in root pulp colour. However, the QTL mapping method mentioned above is extremely dependent on the genetic diversity of the two parents, hence there is variation in QTL effects between populations.30 Genome-wide association studies have been identified to overcome QTL mapping limitations whereby genomic regions responsible for phenotypic traits are identified by using a large size of diverse populations to retrieve significant associations narrowing down the candidate regions.31
Thus, using the genome-wide association studies approach, the same authors, Rabbi et al.18, uncovered two major loci associated with increased pVAC on chromosome 1 at positions 24.1 and 30.5 Mbp. The two SNP-linked markers (S1 24121306 and S1 30543382) explained jointly 81% of total phenotypic variation in root colour score.18 Other studies using independent populations have confirmed the association signals from the same genomic region28,32,33 which contains phytoene synthase 2, a rate-limiting enzyme in the carotenoid biosynthesis pathway34. More recently, five additional QTLs on chromosomes 5 (S5_3387558), 8 (S8 4319215 and S8 25598183), 15 (S15 7659426), and 16 (S16_484011) were identified using a large genome-wide association studies panel of over 5000 cassava accessions.33
Four of the genome-wide uncovered markers (S1 24155522, S1 30543962, S5 3387558, and S8 25598183) linked to pVAC were recently converted to allele-specific PCR assays to facilitate their routine use in MAS.35 We carried out MAS using these SNP markers on a pVAC breeding population developed at the University of Ibadan, Nigeria. We present the performance of the markers in predicting total carotenoid content and root colour score.
Materials and methods
Plant materials and experimentation
The study population was derived from five yellow cassava varieties (IITA-IBA-TMS011368, IITA-IBA-TMS011371, IITA-IBA-TMS011412, IITA-IBA-TMS070593 and IITA-TMS070539) released between 2011 and 2014 and used as female parents in open-pollinated crosses. Before this study, about 3200 half-sib seeds were generated and 89%, 62%, and 0.05% of the progenies were successively screened for different attributes such as cassava mosaic disease severity, plant architecture and root yield among other traits in seedling nursery, clonal evaluation, and preliminary yield stages, respectively.23
Sixty-five (65) cassava genotypes with yellow root tissues selected from preliminary yield trials and two check varieties (IITA-IBA-TMS070593 and IITA-IBA-TMS30555) were evaluated in this study for visual root tissue colour and lab-quantified total carotenoid content (TCC). The experiment was laid out in a randomised complete block design with two replications at the Teaching and Research Farm of the Department of Agronomy, University of Ibadan, Nigeria. To minimise the experimental error due to the relatively large experimental field size, genotypes were divided into two sets. Set 1 and 2 comprised 32 and 33 cassava genotypes, respectively, following the approach of Okechukwu and Dixon36. The check varieties were included in each set. The cassava cuttings were planted at a spacing of 1 m x 1 m with 20 plants representing each genotype in a 20-m2 plot.
Carotenoid assessment in cassava storage roots
The cassava genotypes were selected from preliminary yield trials based on their root colour score. However, to ascertain their actual carotene content, total carotenoid content was measured using iCheck™ (BioAnalyt GmbH, Germany; http://www.bioanalyt.com). The root colour was visually scored using a colour chart with a scale ranging from 1 to 8 where 1 = white; 2 = light cream, 3 = cream, 4 = light yellow, 5 = yellow, 6 = deep yellow, 7 = orange and 8 = pink.37 At harvest (12 months after planting), two to three fully developed roots were selected for TCC quantification using the protocol developed by BioAnalyt laboratory and described by Jaramillo et al.38 The selected cassava roots were properly washed, peeled, and rewashed to remove all impurities. Each root was divided into four vertically; two opposite quarters were chopped into small pieces (to ease the grinding process) and mixed thoroughly. A homogenised sample, 5 g in weight, was collected, macerated using a mortar and pestle, and mixed with 20 mL of distilled water. A final volume of 25 mL was obtained and transferred to a graduated tube. A slurry volume of 0.4 mL was extracted using a syringe and needle and injected into the reagent vial. The vial was allowed to stand for about 5 min to separate the liquid phase before taking and recording the iCheckTM reading.
TCC was calculated as described by Esuma et al.39:

where VS is the total slurry volume (25 mL), WS is the sample weight and Ri stands for the iCheck reading. The iCheck™ carotene analysis procedure was conducted at the Crop Utilisation Laboratory of the Department of Agronomy, University of Ibadan.
DNA extraction and KASP genotyping
The DNA was extracted from young freeze-dried cassava leaves following a modified Dellaporta et al.40 procedure. The DNA quantity and quality were checked using a Nanodrop photometer (ND-8000, Thermo Fisher Scientific, USA) and DNA bands were viewed on agarose gel (1%). The KASP genotyping was performed as described by Codjia et al.41 Four top SNPs (S1 24155522, S1 30543962, S5 3387558 and S8_25598183) coming from phytoene synthase 2 on chromosome 1 and new loci on chromosomes 1, 5 and 8 extracted from Rabbi et al.33 were used for screening the cassava genotypes for pVAC. The pVAC-linked markers successfully converted to allele-specific PCR assay35 enabled the molecular screening. Yellow variety IITA-IBA-TMS 07/0593 (one of the female parents) and white variety IITA-IBA-TMS 30555 were used as positive and negative controls, respectively, in the screening for carotene content. The KASP genotyping was conducted at the International Institute of Tropical Agriculture (IITA), Ibadan.
Statistical analysis
Data on TCC and root colour score were subjected to an analysis of variance (ANOVA) using the Agricolae42 package in R software.43 For the marker-trait association, the best linear unbiased estimator (BLUE) values for TCC and root colour were extracted from a fitted mixed model where blocks and replications were treated as random effects and genotypes, including controls, were regarded as fixed following an adapted mathematical formula for randomised complete block design from Dixon44. Polymorphic information content (PIC) and favourable allele frequencies were calculated using Tassel45 and R software jointly. The markers' effect on TCC and root colour was visually assessed using the ggpubr package.46
For marker predictive ability assessment, a multiple linear regression was computed using the lm function from the lm4 package.47 The traits (TCC or root colour) were considered as response variables and the markers as independent variables (predictors) according to the formula adapted from Uyanik and Güler48:
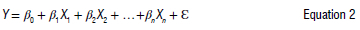
where Y = the predicted value of the trait by the markers, β0 = the constant intercept, β1, β2, ... βn = regression coefficient for each marker, X1,X2 ... Xn = explanatory variables (markers) and ε = residual value.
The best regression model was chosen through a stepwise regression method (both backward and forward) using the Mass package. For model validation, a bootstrapped sampling strategy was applied (n = 10) using the tidymodels package. The correlation matrix plot was performed using the corrplot package.49
Results and discussion
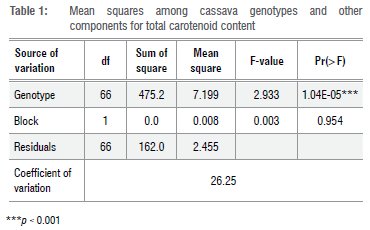
Variation for TCC among cassava genotypes
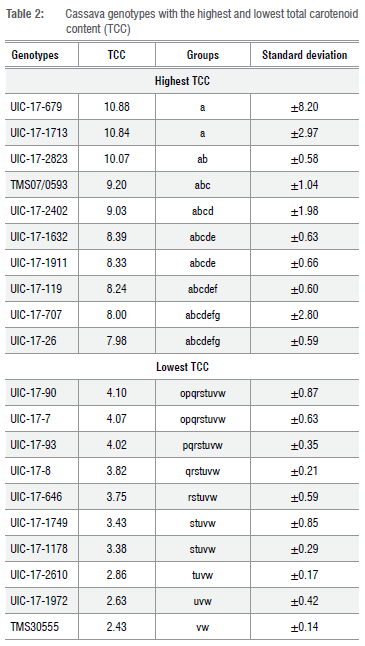
The analysis of variance revealed highly significant differences among the yellow cassava genotypes for TCC, but no significant difference among block components was observed (Table 1). The coefficient of variation (CV) was 26%. Genotypes UIC-17-679, UIC-17-1713, and UIC-17-2823 had TCCs ranging from 10.07 μg/g to 10.88 μg/g. These values are higher than the TCC value of 9.20 μg/g recorded in the yellow check variety IITA-TMS-070593 (Table 2).
Frequency distribution and correlation between TCC and root colour score
Data on TCC followed a normal distribution (Figure 1). The TCC ranged from 2.43 μg/g to 10.88 μg/g with a mean of 5.88 μg/g. The highest TCC recorded in this study (10.88 μg/g) is similar to the maximum TCC (11 μg/g) reported by Esuma et al.39 among the F1 progenies of cassava using the iCheckTM carotene determination procedure. The root colour ranged from 1 to 4 with a mean of 3 in the panel of yellow cassava genotypes (Figure 1).
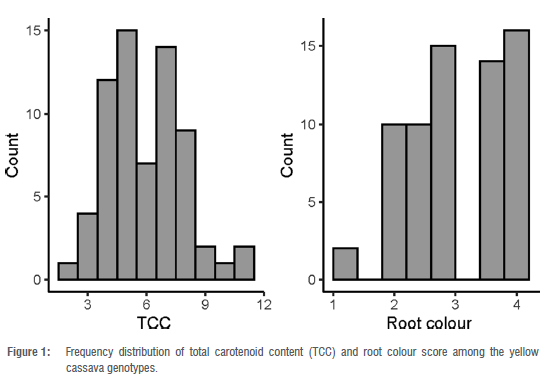
A highly significant correlation (r = 0.88; p < 0.0001) was observed between visual root colour score and TCC using iCheckTM (Figure 2), which is consistent with earlier submissions.13,50,51 Screening large cassava populations becomes much more motivating due to the existing correlation between TCC and root colour score.39 However, lab-based quantification of carotenoid content is still needed at later stages of selection in order to have more precise estimates, for example when recommending new genotypes for official registration and release.52
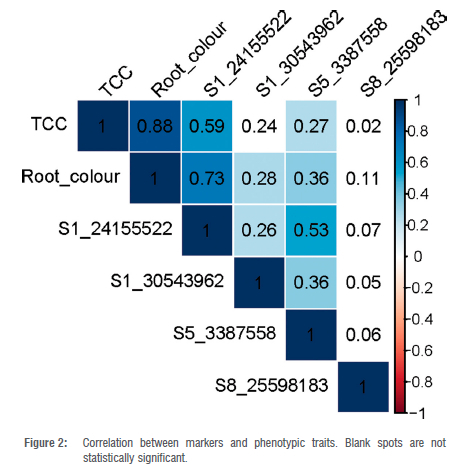
Marker informativeness, technical metrics and genotypic population structure
The favourable allele frequencies for the SNP markers varied from 0.20 to 0.87 with an average of 0.49 (Table 3). The observed heterozygosity at the studied loci ranged from 0.19 to 0.89 with a mean of 0.51. The polymorphic information content (PIC) - which is a crucial metric for the informativeness of a marker - varied from 0.22 (marker S1 24155522) to 0.49 (marker S5 3387558) with an average of 0.38 (Table 3). The SNP markers S1 24155522, S1 30543962, S5 3387558, and S8 25598183 had call rates of 94%, 98.5%, 98.5% and 97%, respectively.
Approximately 90% of the study population were identified with favourable 'A' allele at marker S1 24155522 while about 3% of the genotypes did not have the favourable allele (Table 4). Furthermore, 72% were homozygotes fixed for the useful allele A whereas about 18% were found heterozygotes (CA) at phytoene synthase linked marker (Table 4).
Effects of favourable and unfavourable alleles on TCC and root tissue colour
Exploratory analysis of genotype classes (AA, CA, CC) at SNP S1 24155522 versus TCC revealed a highly significant difference (p < 0.0001) (Figure 3). The individuals with two copies of favourable alleles (AA) at phytoene synthase locus had a TCC mean of 6.52 μg/g, whereas most of the genotypes (CC) with the unfavourable allele had an average TCC of 2.54 μg/g. The majority of heterozygous genotypes (CA) had an intermediate level of TCC (4.25 μg/g). A f-test (pairwise comparison) revealed a significant difference at the 5% level at the other loci, except for marker S8_25598183 which was not significant in discriminating between genotypic classes for TCC trait (Figure 3). Similar observations were made for root colour scores at all loci, excluding S8 25598183 (Figure 4).
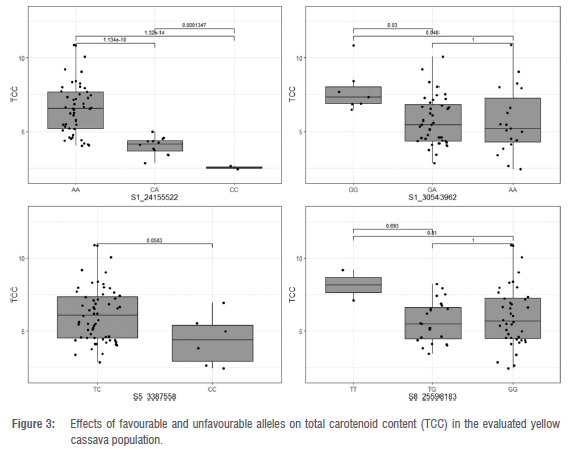
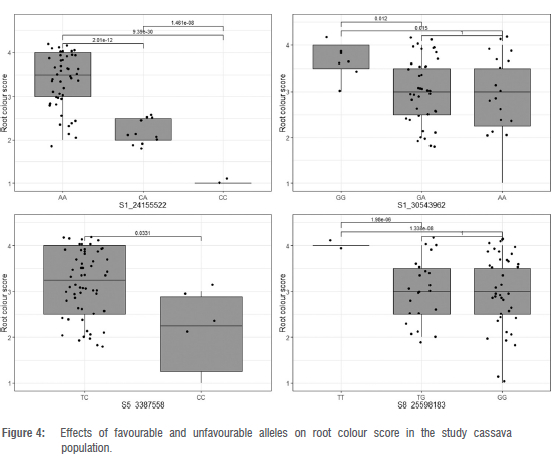
In a recent study conducted by Udoh et al.13, the marker at PSY2 gene (S1_24155522) was significantly associated with root colour score and TCC. Phytoene synthase had previously been identified as a gene engaged in the initial step of cassava carotenoid biosynthesis.18,34 Phytoene synthase is known for the accumulation of provitamin A carotenoid in cassava roots.28,34 This study confirms the primary role of the PSY enzyme (Manes.01G124200), which converts geranylgeranyl diphosphate to phytoene.18,34 Beta-carotene constitutes the main component of TCC in cassava17,53, suggesting that the products of PSY2 are mostly channelled towards the lycopene beta cyclase (lcyB) part of the pathway.54 Further research is needed to find markers that select for down-regulation of the activity of lycopene epsilon cyclase (lcyE), which will even boost the proportion of beta-carotene.55
Carotenoid accumulation in cassava is driven by an additive gene effect.28 This observation is supported by the differential allele effect observed in the box-plot analysis for marker PSY_572/ S1 24155522 (Figures 3 and 4). Keller et al.56 defined the additive effect as an allele substitution's effect at a locus that can explain half of the difference of homozygous individuals' performance as observed in this study at PSY2 gene (Figures 3 and 4). The additive nature of the provitamin A trait suggests that the accumulation of TCC in cassava storage roots would be improved through a recurrent selection allowing gene recombination within the breeding set.16,28,51
Prediction of TCC and root colour by markers using multiple linear regression model
Individually, the marker at the phytoene synthase gene explained much of the phenotypic variation in TCC (R2 = 0.37) while the other markers -S1 30543962, S5 3387558 and S8 25598183 - explained 11%, 7% and 5%, respectively. For root colour, each marker S1 24155522, S1 30543962, S53387558 and S8 25598183 accounted separately for 55%, 8%, 16% and 5%, respectively.
For markers predictive ability assessment, a multiple linear regression was computed with the four markers which explained 39% of the total variation in TCC in the present population (adjusted R2=0.39; p = 2.78E-05) (Figure 5). On the other hand, these SNP markers together explained up to 54% of the total variation in visual root colour (adjusted R2=0.54; p = 3.901E-08) (Figure 5). However, the best model chosen using a stepwise regression method returned a simple model only fitted with the most significant predictor at the phytoene synthase gene indicating the elimination of other non-significant markers in the prediction model for TCC and root colour. The low phenotypic variation of TCC and root colour explained by the three markers S1 30543962, S5 3387558 and S8 25598183 suggests that the newly uncovered QTLs for pVAC have a minor effect on the trait, as previously reported by Ige et al.35
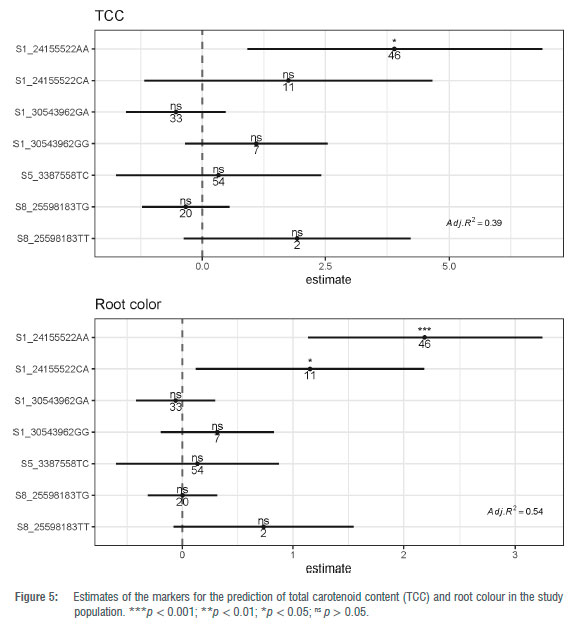
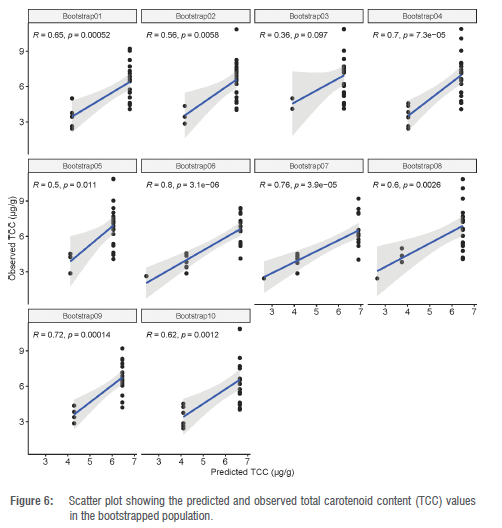
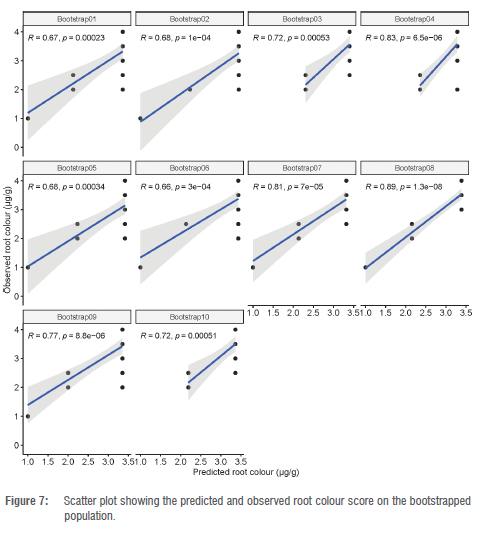
The best prediction model fitted with the most significant marker was validated using a bootstrap resampling strategy. The correlation coefficient between the predicted and the observed TCC values ranged from 0.36 to 0.80 with an average of 0.63 (Figure 6) whereas a correlation ranging from 0.66 to 0.89 with a mean of 0.75 was found between predicted and observed root colour score (Figure 7). These results confirm that marker S1 24155522 has sufficient predictive ability for usage in marker-assisted selection to enhance provitamin A content in African cassava storage roots.
Phenotypic and molecular improvement for provitamin A content
In this study, the maximum TCC recorded in the cassava population was 10.88 μg/g - a value that, although higher than that of the national yellow control variety, still does not reach the breeding target (15 μg/g). The rapid-cycling recurrent selection approach is known to increase carotenoid accumulation in cassava storage roots, especially in breeding programmes targeting exclusively carotenoid content enhancement.16 However, the method relies on precise carotenoid quantification using laboratory methods such as high-performance liquid chromatography (HPLC) which is costly, time consuming and labour intensive.16,21 The use of SNP markers is expected to facilitate rapid population improvement and overcome the challenges associated with phenotype-based selection.57
In our study, we used iCheck to quantify total carotenoid content rather than individual carotenoid components (alpha-carotene, all-trans beta-carotene, violaxanthin, lutein, 15-cis beta-carotene, 13-cis beta-carotene, 9-cis beta-carotene, and phytoene), a process that would require HPLC. In cassava, it is well established that more than 80% of TCC is total beta-carotene32, a precursor of vitamin A known to be readily converted into retinol by the body58 . Additionally, various studies have reported a high positive correlation between TCC and total beta-carotene in cassava storage roots.16,32,59
The accuracy of MAS found in this study for root colour score is 55%. A similar study using different cassava populations resulted in a higher accuracy of 80%.35 The relatively lower prediction accuracy from our study could be explained by the small population size derived from five parents and the narrow range of variation in root pigmentation (between cream and yellow) after discarding white root clones at the early stages of selection.
Nevertheless, the observed accuracy from relatively few markers is in agreement with previous studies which have shown simple inheritance of carotenoid traits in cassava.51,60 Further, the accuracy obtained from the present study is similar to that obtained from genome-wide predictions.32,61 Ikeogu et al.32 reported prediction accuracies up to 52% in a genomic selection with 594 cassava genotypes using the multiple trait carotenoids (individual TC components). Equally, Esuma et al.61 reported an equivalent accuracy of 52% in root colour score while predicting carotenoids using genomic tools. This shows that MAS is sufficient for an early screening of individuals for carotenoid content. MAS is much more suitable for traits controlled by a few or major loci62 as observed in this study with the major PSY2 gene known to be responsible for provitamin A accumulation and tagged by marker S1_24155522. The approach is particularly convenient when dealing with traits that are expensive to phenotype, such as carotenoid content, and which can only be evaluated at harvest. We demonstrate the suitability of carotenoid-content-linked SNPs for MAS in our breeding population. The present results open avenues for breeders to more comfortably fast-track the transfer of useful alleles of carotenoid genes into elite cassava breeding lines, thus contributing to the reduction of VAD prevalence, especially among pregnant women and children in developing countries.
Conclusion
We carried out marker-assisted selection to increase provitamin A content in a cassava breeding population using four SNP markers and laboratory-quantified carotenoid content as well as root colour score. The marker accuracy evaluation revealed that the major PSY2 locus on chromosome 1, tagged by S1_24155522 and driven by an additive gene effect, explained a relatively high phenotypic variation in TCC and root colour in the study population and could be deployed for MAS in cassava breeding programmes. The top three genotypes - UIC-17-679, UIC-17-1713, and UIC-17-2823 - with TCC greater than that of the national yellow check IITA-IBA-TMS070593 are recommended to be advanced to the next breeding stages for evaluation of other key traits such as root yield, dry matter, cyanide content, and root mealiness across multi-environments to satisfy farmers' and consumers' needs.
Acknowledgements
We thank the cassava improvement programme of the University of Ibadan, Nigeria, for providing the plant material and the carotene analysis facilities. We acknowledge support from the International Institute of Tropical Agriculture for the molecular genotyping facilities. E.D.C. is indebted to the African Union Commission for a PhD scholarship in plant breeding at PAULESI, Ibadan, Nigeria. We appreciate financial support from the African Union Commission.
Competing interests
We have no competing interests to declare.
Authors' contributions
E.D.C.: Conceptualisation; methodology; data collection; sample analysis; data analysis; data curation; writing - the initial draft; funding acquisition. B.O.: Conceptualisation; methodology; data curation; writing -revisions; student supervision; project management; development of the cassava population. I.Y.R.: Conceptualisation; methodology; writing -revisions; student supervision; project management; development of the molecular markers. C.E.U.: Data analysis.
References
1. Pelletier DL, Frongillo EA, Schroeder DG, Habicht JP The effects of malnutrition on child mortality in developing countries. Bull World Health Organ. 1995;73(4):443-448. [ Links ]
2. Ecker O, Nene M. Nutrition policies in developing countries: Challenges and highlights. Washington: International Food Policy Research Institute; 2012. https://agritech.tnau.ac.in/nutrition/nutritionpolicies_pn.pdf [ Links ]
3. Nestel P, Bouis H, Meenakshi J, Pfeiffer W. Biofortification of stable food crops. J Nutr. 2006;136:1064-1067. https://doi.org/10.1093/jn/136.4_1064 [ Links ]
4. Beyene G, Solomon FR, Chauhan RD, Gaitán-Solis E, Narayanan N, Gehan J, et al. Provitamin A biofortification of cassava enhances shelf life but reduces dry matter content of storage roots due to altered carbon partitioning into starch. Plant Biotechnol J. 2018;16:1186-1200. https://doi.org/10.1111/pbi.12862 [ Links ]
5. World Health Organization (WHO). Nutrition health topics. Rome: WHO; 2022. https://www.who.int/data/nutrition/nlis/info/vitamin-a-deficiency [ Links ]
6. Maziya-Dixon B, Akinyele IO, Oguntona EB, Nokoe S, Sanusi RA, Harris E. Nigeria food consumption and nutrition survey, 2001-2003 summary. Ibadan: International Institute of Tropical Agriculture; 2004. [ Links ]
7. Zhu C, Naqvi S, Gomez-Galera S, Pelacho AM, Capell T, Christou P Transgenic strategies for the nutritional enhancement of plants. Trends Plant Sci. 2007;12(12):548-555. https://doi.org/10.1016/j.tplants.2007.09.007 [ Links ]
8. Lebot V. Tropical root and tuber crops: Cassava, sweet potato, yams, and aroids. In: Atherton J, Rees A, editors. Crop production science in horticulture series. Vol 17. Wallingford: CAB International; 2008. p. 1-433. https://books.google.com.ng/books?id=oUAcKBhfiOEC&printsec=frontcover&hl=fr&source=gbs_atb#v=onepage&q&f=false [ Links ]
9. Dos Santos CS, Sousa MB, Brito AC, De Oliveira LA, Carvalho CWP De Oliveira EJ. Genome-wide association study of cassava starch paste properties. PLoS One. 2022;17(1):1-26. https://doi.org/10.1371/journal.pone.0262888 [ Links ]
10. International Institute of Tropical Agriculture (IITA). Cassava in tropical Africa, a reference manual. Ibadan: IITA; 1990. https://www.iita.org/wp-content/uploads/2016/06/Cassava_in_tropical_Africa_a_reference_manual_1990.pdf [ Links ]
11. Sanyang S, Ryburn R, Mur R. Against the grain and to the roots. In: Audet-Bélanger G, Dia A, editors. Arnhem: LM Publishers; 2014. https://www.kit.nl/publication/against-the-grain-and-to-the-roots/ [ Links ]
12. Okwuonu IC, Narayanan NN, Egesi CN, Taylor NJ. Opportunities and challenges for biofortification of cassava to address iron and zinc deficiency in Nigeria. Glob Food Sec. 2021;28:100478. https://doi.org/10.1016/j.gfs.2020.100478 [ Links ]
13. Udoh LI, Gedil M, Parkes EX Kulakow F; Adesoye, A, Nwuba C, et al. Candidate gene sequencing and validation of SNP markers linked to carotenoid content in cassava (Manihot esculenta Crantz). Mol Breed. 2017;37(10):1-12. https://doi.org/10.1007/s11032-017-0718-5 [ Links ]
14. Muthayya S, Rah JH, Sugimoto JD, Roos FF, Kraemer K, Black RE. The global hidden hunger indices and maps: an advocacy tool for action. PLoS One. 2013;8(6):1-12. https://doi.org/10.1371/journal.pone.0067860 [ Links ]
15. Ruel-Bergeron JC, Stevens GA, Sugimoto JD, Roos FF, Ezzati M, Black RE, et al. Global update and trends of hidden hunger, 1995-2011: The hidden hunger Index. PLoS One. 2015;10(12):1-13. https://doi.org/10.1371/journal.pone.0143497 [ Links ]
16. Ceballos H, Morante N, Sánchez T Ortiz D, Aragón I, Chávez AL, et al. Rapid cycling recurrent selection for increased carotenoids content in cassava roots. Crop Sci. 2013;53:2342-2351. https://doi.org/10.2135/cropsci2013.02.0123 [ Links ]
17. Sanchez T, Ceballos H, Dufour D, Ortiz N, Morante F, Calle T, et al. Prediction of carotenoids, cyanide and dry matter contents in fresh cassava root using NIRS and Hunter color techniques. Food Chem. 2014;151:444-451. https://doi.org/10.1016/j.foodchem.2013.11.081 [ Links ]
18. Rabbi IY Udoh LI, Wolfe M, Parkes EY Gedil MA, Dixon A, et al. Genome-wide association mapping of correlated traits in cassava: Dry matter and total carotenoid content. Crop Sci Soc Am. 2017;10(3):1-14. https://doi.org/10.3835/plantgenome2016.09.0094 [ Links ]
19. Ilona IP Bouis H, Palenberg M, Moursi M, Oparinde A. Vitamin A cassava in Nigeria: Crop development and delivery. Afr J Food Agric Nutr Dev. 2017;17(02):12000-12025. https://doi.org/10.18697/ajfand.78.HarvestPlus09 [ Links ]
20. De Moura FF, Moursi M, Lubowa A, Ha B, Boy E, Oguntona B, et al. Cassava intake and Vitamin A status among women and preschool children in Akwa-Ibom, Nigeria. PLoS One. 2015;10(6):1-14. https://doi.org/10.1371/journal.pone.0129436 [ Links ]
21. Andersson MS, Saltzman A, Virk PS, Pfeiffer WH. Progress update: Crop development of biofortified staple food crops under HarvestPlus. Afr J Food Agric Nutr Dev. 2017;17(2):11905-11935. https://doi.org/10.18697/ajfand.78.HarvestPlus05 [ Links ]
22. Collard BCY Mackill DJ. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B Biol Sci. 2007;363(1491):557-572. https://doi.org/10.1098/rstb.2007.2170 [ Links ]
23. Olasanmi B, Kyallo M, Yao N. Marker-assisted selection complements phenotypic screening at seedling stage to identify cassava mosaic disease-resistant genotypes in African cassava populations. Sci Rep. 2021;11(1):1-8. https://doi.org/10.1038/s41598-021-82360-8 [ Links ]
24. Staub JE, Serquen FC, McCreight JD. Genetic diversity in cucumber (Cucumis safivus L.): III. An evaluation of indian germplasm. Genet Resour Crop Evol. 1997;44(4):315-326. https://doi.org/10.1023/A:1008639103328 [ Links ]
25. Jonah PM, Bello LL, Lucky O, Midau A, Moruppa SM. Review: The importance of molecular markers in plant breeding programmes. Glob J Sci Front Res. 2011;11(5):9. [ Links ]
26. Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34:275-305. https://doi.org/10.1051/gse:2002009 [ Links ]
27. Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol Breed. 2014;33(1):1-14. https://doi.org/10.1007/s11032-013-9917-x [ Links ]
28. Esuma W, Herselman L, Labuschagne MT, Ramu P Lu F, Baguma Y et al. Genome-wide association mapping of provitamin A carotenoid content in cassava. Euphytica. 2016;212(1):97-110. https://doi.org/10.1007/s10681-016-1772-5 [ Links ]
29. Rabbi I, Hamblin M, Gedil M, Kulakow P Ferguson M, Ikpan AS, et al. Genetic mapping using genotyping-by-sequencing in the clonally propagated cassava. Crop Sci. 2014;54:1384-1396. https://doi.org/10.2135/cropsci2013.07.0482 [ Links ]
30. Borevitz JO, Nordborg M. The impact of genomics on the study of natural variation in arabidopsis. Plant Physiol. 2003;132:718-725. https://doi.org/10.1104/pp.103.023549 [ Links ]
31. Korte A, Ashley F. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods. 2013;9:29. https://doi.org/10.1186/1746-4811-9-29 [ Links ]
32. Ikeogu UN, Akdemir D, Wolfe MD, Okeke UG, Chinedozi A, Jannink J-L, et al. Genetic correlation, genome-wide association and genomic prediction of portable NIRS predicted carotenoids in cassava roots. Front Plant Sci. 2019;10:1-11. https://doi.org/10.3389/fpls.2019.01570 [ Links ]
33. Rabbi IY Kayondo S, Bauchet G, Yusuf M, Aghogho CI, Ogunpaimo K, et al. Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol Biol. 2022;109:195-213. https://doi.org/10.1007/s11103-020-01038-3 [ Links ]
34. Welsch R, Arango J, Bar C, Salazar B, Al-Babili S, Beltrán J, et al. Provitamin A accumulation in cassava (Manihof esculenfa) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell. 2010;22:3348-3356. https://doi.org/10.1105/tpc.110.077560 [ Links ]
35. Ige A, Olasanmi B, Bauchet GJ, Kayondo IS, Mbanjo EGN, Uwugiaren R, et al. Validation of SNP markers for marker-assisted selection of genotypes with increased carotenoid and dry matter contents in cassava. Front Plant Sci. 2022;13:1-17. https://doi.org/10.3389/fpls.2022.1016170 [ Links ]
36. Okechukwu RU, Dixon AGOO. Genetic gains from 30 years of cassava breeding in Nigeria for storage root yield and disease resistance in elite cassava genotypes. J Crop Improv. 2008;22(2):181-208. https://doi.org/10.1080/15427520802212506 [ Links ]
37. Okoro M, Egesi C, Olasanmi B, Chimaobi I, Njoku S, Ikeogu U, et al. Clonal evaluation trial of yellow root cassava genotypes in south eastern Nigeria. Paper presented at: Second Conference of the Global Cassava Partnership; 2012 June 18-22; Kampala, Uganda. [ Links ]
38. Jaramillo A, Londono L, Orozco J, Patino G, Belalcazar J, Davrieux F. A comparison study of five different methods to measure carotenoids in biofortified yellow cassava (Manihot esculenta). PLoS One. 2018;13(12):1-14. https://doi.org/10.1371/journal.pone.0209702 [ Links ]
39. Esuma W, Kawuki RS, Herselman L, Labuschagne MT. Diallel analysis of provitamin A carotenoid and dry matter content in cassava (Manihot esculenta Crantz). Breed Sci. 2016;66(4):627-635. https://doi.org/10.1270/jsbbs.15159 [ Links ]
40. Dellaporta SL, Wood J, Hicks JB. A plant DNA mini preparation: Version II. Plant Mol Biol Report. 1983;1(4):19-21. https://doi.org/10.1007/BF02712670 [ Links ]
41. Codjia ED, Olasanmi B, Agre PA, Uwugiaren R, Ige AD, Rabbi IY Selection for resistance to cassava mosaic disease in African cassava germplasm using single nucleotide polymorphism markers. S Afr J Sci. 2022;118(1/2), Art. #11607. https://doi.org/10.17159/sajs.2022/11607 [ Links ]
42. Mendiburu F. Agricolae: Statistical procedures for agricultural research. R Package Version 1.2-3. Available from: http://CRAN.R-project.org/package=agricolae. [ Links ]
43. R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2020 http://www.r-project.org/index.html [ Links ]
44. Dixon P Should blocks be fixed or random? In: Proceedings of the Conference on Applied Statistics in Agriculture. New Priarie Press; 2016. p. 23-39. https://doi.org/10.4148/2475-7772.1474 [ Links ]
45. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633-2635. https://doi.org/10.1093/bioinformatics/btm308 [ Links ]
46. Kassambara. Ggpubr "ggplot2" based publication ready plots. 2023. Available from: https://github.com/kassambara/ggpubr. [ Links ]
47. Bates D, Mâchler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1-48. https://doi.org/10.18637/jss.v067.i01 [ Links ]
48. Uyanik GK, Güler N. A study on multiple linear regression analysis. Procedia - Soc Behav Sci. 2013;106:234-240. https://doi.org/10.1016/j.sbspro.2013.12.027 [ Links ]
49. Wei T, Simko V Wei T, Simko V. R package "corrplot": Visualization of a correlation matrix. Version 0.92. 2021. Available from: https://github.com/taiyun/corrplot. [ Links ]
50. Sanchez T, Chávez AL, Ceballos H, Rodriguez-Amaya DB, Nestel P Ishitani M. Reduction or delay of post-harvest physiological deterioration in cassava roots with higher carotenoid content. J Sci Food Agric. 2006;86(4):634-639. https://doi.org/10.1002/jsfa.2371 [ Links ]
51. Akinwale MG, Aladesanwa RD, Akinyele BO, Dixon AGO, Odiyi AC. Inheritance of β-carotene in cassava (Manihot esculenta Crantz). Int J Genet Mol Biol. 2010;2(10):198-201. [ Links ]
52. Iglesias C, Mayer J, Chavez L, Calle F. Genetic potential and stability of carotene content in cassava roots. Euphytica. 1997;94:367-373. https://doi.org/10.1023/A:1002962108315 [ Links ]
53. Carvalho LJCB, Agustini MAV, Anderson JV, Vieira EA, De Souza CRB, Chen S. Natural variation in expression of genes associated with carotenoid biosynthesis and accumulation in cassava (Manihot esculenta Crantz) storage root. BMC Plant Biol. 2016;16(1):1-23. https://doi.org/10.1186/s12870-016-0826-0 [ Links ]
54. Apel W, Bock R. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin a conversion. Plant Physiol. 2009;151(1):59-66. https://doi.org/10.1104/pp.109.140533 [ Links ]
55. Ceballos H, Davrieux F, Talsma EF, Belalcazar J, Chavarriaga PS. Carotenoids in cassava roots. Carotenoids. 2017:189-221. https://doi.org/10.5772/intechopen.68279 [ Links ]
56. Keller B, Feuillet C, Messmer M. Genetics of disease resistance. In: Slusarenko A, Fraser RS, Van Loon LC, editors. Mechanisms of resistance to plant diseases. Dordrecht: Springer; 2000. p. 101-160. https://doi.org/10.1007/978-94-011-3937-3_5 [ Links ]
57. Kebede D, Mengesha W, Menkir A, Abe A, Garcia-Oliveira AL, Gedil M. Marker based enrichment of provitamin A content in two tropical maize synthetics. Sci Rep. 2021;11:1-10. https://doi.org/10.1038/s41598-021-94586-7 [ Links ]
58. Greenwald P McDonald S. The β-carotene story. In: Advances in experimental medicine and biology. Vol 492. Boston, MA: Springer; 2001. p. 219-231. https://doi.org/10.1007/978-1-4615-1283-7_17 [ Links ]
59. Tufchi RM, Shastry M, Singh NK. Phenotyping of maize inbred lines for beta-carotene and determining relationship with total carotenoids and kernel colour in maize. Indian J Genet Plant Breed. 2014;74(4):631-637. https://doi.org/10.5958/0975-6906.2014.00902.X [ Links ]
60. Chavez AL, Bedoya JM, Sanchez T, Iglesias C, Ceballos H, Roca W. Iron, carotene, and ascorbic acid in cassava roots and leaves. Food Nutr Bull. 2000;21(4):410-413. https://doi.org/10.1177/156482650002100413 [ Links ]
61. Esuma W, Ozimati A, Kulakow P, Gore MA, Wolfe MD, Nuwamanya E, et al. Effectiveness of genomic selection for improving provitamin A carotenoid content and associated traits in cassava. G3 Genes Genom Genet. 2021;11(9):1-10. https://doi.org/10.1093/g3journal/jkab160 [ Links ]
62. Mbanjo EGN, Rabbi IY, Ferguson ME, Kayondo SE, Eng NW, Tripathi L, et al. Technological innovations for improving cassava production in sub-Saharan Africa. Front Genet. 2021;11. https://doi.org/10.3389/fgene.2020.623736 [ Links ]
 Correspondence:
Correspondence:
Ismail Rabbi
Email: i.rabbi@cgiar.org
Received: 04 Nov. 2022
Revised: 08 July 2023
Accepted: 17 July 2023
Published: 29 Nov. 2023
Editor: Teresa Coutinho
Funding: African Union Commission














