Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.119 no.9-10 Pretoria Set./Out. 2023
http://dx.doi.org/10.17159/sajs.2023/10901
RESEARCH ARTICLE
Accessory gene regulators and virulence genes associated with the pathogenicity of Staphylococcus aureus from clinical and community settings in Lagos, Nigeria
Nkechi V. EnwuruI; Solayide A. AdesidaII; Christian A. EnwuruIII; Udoma E. MendieI
IDepartment of Pharmaceutical Microbiology and Biotechnology, Faculty of Pharmacy, College of Medicine, University of Lagos, Lagos, Nigeria
IIDepartment of Microbiology, Faculty of Science, University of Lagos, Lagos, Nigeria
IIICentre for Infectious Disease Research, Microbiology Department, Nigerian Institute of Medical Research, Lagos, Nigeria
ABSTRACT
Staphylococcus aureus is a prominent pathogen that causes serious community and hospital-acquired infections globally. Its pathogenicity is attributed to a variety of secreted and cell surface associated proteins that are modulated by the quorum-sensing accessory gene regulator (agr) system. In this study, we investigated the presence of toxin genes and agr involved with S. aureus from clinical samples and apparently healthy individuals. Unequivocal identification of the isolates was obtained with the Vitek 2 system. We screened 70 clinical (CL) and 22 community (C) S. aureus strains for the methicillin resistance (mecA) gene, agr and superantigens (SAg) (enterotoxins and toxic shock syndrome toxin-1) using PCR techniques. A total of 12 clinical isolates were classified as methicillin-resistant S. aureus (MRSA); 89 isolates belonged to one of the four agr groups (agr14), and 3 isolates were non-typeable. Of the agr groups, agr1 was the most prominent and mostly consisted of isolates from pus/wounds. The methicillin-susceptible S. aureus (MSSA) isolates were distributed within the four agr groups while MRSA strains were restricted to agr1 and agr3. The most common enterotoxin gene, sei, was likewise more prevalent in MSSA strains than in MRSA strains, where sea predominated. The co-existence of two or more enterotoxins was confirmed in 40% of the isolates. sea occurred through all the agr groups except agr3 and sei was not found in agr1 and agr4. The toxic shock toxin (tst) gene was detected in six MSSA. These findings suggest that MSSA may cause more lethal infections than MRSA because of the increased frequency of toxic genotypes seen in MSSA strains.
SIGNIFICANCE:
• Isolates in the agr1,3 groups had more SAg toxin genes, whereas isolates in the agr4 groups possessed more tst genes.
• The MSSA isolates contained higher proportions of virulence genes than MRSA.
• The clinical implications of this discovery include that MSSA may cause more lethal infections than MRSA due to the greater number of toxigenic genotypes discovered.
Keywords: accessory gene regulators, pathogenesis, Staphylococcus aureus, superantigens
Introduction
Staphylococcus aureus is a dynamic Gram-positive pathogen that lives as a harmless commensal bacterium on the skin and mucosal surfaces of humans and other animals.1 It has the potential to multiply in the blood and other tissues, triggering serious medical conditions.2 It is widely considered as one of the leading causes of hospital- and community-acquired infections globally.3 Evidently, the organism features prominently in 8-33% of cases of skin, soft-tissue, and bloodstream infections that can result in significant morbidity and mortality.4
Depending on its growth phases, S. aureus is able to utilise a wide range of virulence factors to initiate and establish infections in susceptible hosts. Typically, in the lag and early exponential growth phrases, the pathogen releases cell-wall-associated factors that aid in tissue adhesion and immune system evasion.5 When the bacterial population gets to the late exponential growth phase, it begins to secrete a wide range of exoproteins, including proteases, haemolysins, and superantigens (SAgs) while also down-regulating cell-wall-associated factors, resulting in biofilm dispersion and dissemination of infection.5
Staphylococcal superantigens (SAgs) are notable exotoxins which play a critical role in S. aureus infections. They have been categorised into staphylococcal enterotoxins (SE), staphylococcal enterotoxin-Wre (SEl) proteins and toxic-shock syndrome toxin6 encoded by the tst gene. The adhesion and invasion phases of S. aureus development are characterised pre-eminently by its population-density-dependent behaviour. The synchronisation and swift transition between these two phases is achieved through a cell-to-cell communication mechanism known as quorum sensing (QS). The majority of these virulence genes are regulated by the accessory gene regulator (agr) system which is divided into four (1-4) agr groups.7,8
The agr region is crucial in pathogenesis and actively controls the expression of virulence factors, heterogeneous resistance in methicillin-resistant S. aureus (MRSA), and biofilm development.911 It co-regulates the expression of several exoproteins including a-, β-, y-haemolysin as well as lipases, phenol-soluble modulins and TSST-1 while down regulating the synthesis of cell-wall-associated proteins such as protein A, coagulase, and fibronectin binding protein.12,13 Several studies have been conducted to investigate the association of agr groups with certain biological traits in S. aureus and the outcomes have indicated that some enterotoxin clusters - namely seg, sei, sem, sen, and seo - are linked to agr4 in a number of strains.14
In Tehran, Choopani et al.15 noticed that MRSA isolates had the seb gene but that the TSST-1 precursor antigen was predominantly expressed by agr-III and IV strains. A study Some studies in southwest Nigeria indicated that 89% of the isolates screened had at least one SE: seo (34%) was the most prominent SE, then seg (30%) and sea (21%), whereas toxic shock syndrome toxin (TSST), seb, sec, see, sej, sel, sem, ser and seu were not detected. Similarly, the co-existence of seo/ seg and sei/seg genes were recognised.16
Also, Akinduti et al.17 reported that the bulk of the enterotoxins detected among clinical methicillin-sensitive S. aureus isolates in their study were confined to agr2. Nonetheless, information on the extent of superantigen genetic heterogeneity in community and clinical S. aureus populations in Nigeria is limited. Consequently, in the present study, we focused on determining the presence of staphylococcal superantigen genes in S. aureus isolates from two healthcare institutions and apparently healthy volunteers. Co-existence of the toxigenic genes with the agr groups and sources of the isolates was also assessed.
Materials and methods
Bacterial isolates
We analysed 92 previously described non-duplicated S. aureus strains collected over a period of 2 years from two tertiary care hospitals (Lagos University Teaching Hospital (LUTH), Idi-araba and National Orthopaedic Hospital, Igbobi).18 The sample consisted of 22 nasal (NS) isolates obtained from apparently healthy individuals and designated as community (C) strains and 70 isolates obtained from various specimens submitted to the microbiological laboratories of the hospitals, classified as the clinical (CL) strains.
Bacterial identification and methicillin resistance determination
The isolates were identified using Vitek 2 automated systems (BioMérieux, Marcy L'Étoile, France). Antibiotic susceptibility testing of the isolates, also carried out by the Vitek 2 automated system, had been determined previously.18
Genomic DNA extraction
Genomic DNA for the evaluation of mecA, enterotoxin, toxic shock syndrome toxin-1 genes and agr determinants was extracted as previously described.18 The quality and concentration of DNA were estimated spectrophotometrically.
mecA PCR
Strains that demonstrated phenotypic resistance to methicillin were subjected to mecA PCR as previously described.19 The PCR amplification was carried out in a 25 reaction volume containing 1 of template DNA, 10 nL of 2 x master mix of 1 x PCR buffer, 1.5 mmol/L MgCl2, 0.15 mmol/L dNTP and 1.25 IU Taq DNA polymerase, 0.7 of 0.8 nmol/L of each primer and 12.6 of deionised water. For the amplification of the 533 base pair (bp) fragment, the mecA-specific primer pairs employed were MECAP4: 5' - TCCAGATTACAACTTCACCAGG - 3', and MECAP7: 5' - CCACTTCATATCTTGTAACG - 3'. The PCR products were then separated by agarose gel electrophoresis and visualised with ethidium bromide staining. A 100 bp dNa ladder was used as a molecular weight marker.
Determination of agr groups
We used the primers and thermal cycling conditions for agr groups differentiation described by Shopsin et al.20 The S. aureus strains RN6390 (agr group I), RN6607 (agr group II), RN8465 (agr group III), and RN4850 (agr group IV), graciously provided by the Medical Microbiology Laboratory, Otto-von-Guericke University, Germany, were used as controls.
Detection of superantigen genes
Multiplex PCR was used to determine enterotoxin genes associated with S. aureus.21 We employed the primers for classical staphylococcal enterotoxin genes (sea, seb, sec and sed) as well as toxic shock syndrome toxin-1 (tst) and the SEl (see, seg, seh, sei, sej). For quality control, S. aureus ATCC 19095 was used for sec, seh, seg and sei while S. aureus FR I913 was employed for sea, see, tst and S. aureus ATCC 14458 for seb gene.
Statistical analyses
The data were analysed using GraphPad Prism 7 software (San Diego, CA, USA) and Microsoft Excel. Data for frequencies in percentages and absolute values are shown in charts and tables.
Ethical approval
The study was approved by the Institutional Review Board (IRB) of the College of Medicine, University of Lagos, Nigeria (reference number: CM/COM/8/VOL.XIX).
Results
Staphylococcus aureus distribution and methicillin resistance
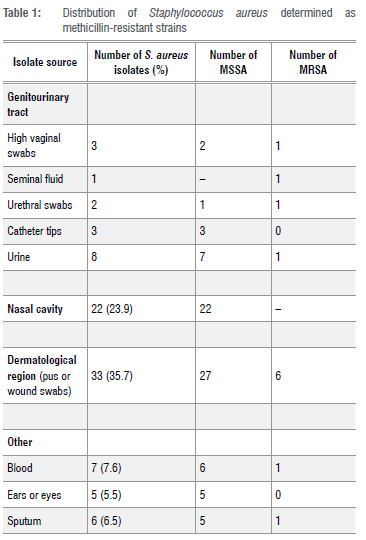
The S. aureus isolates investigated in this study originated mainly from three body sites: the genitourinary tract, nasal cavity and dermatological region of the body. Additionally, isolates were obtained from sputum (4.3%), blood (7.6%), and the ear or eye (6, 6.5%). Twelve clinical isolates were identified as MRSA and harboured the mecA gene (Table 1).
Classification of agr alleles
With the exception of three isolates that were non-typeable, the S. aureus isolates were classified into four groups using the agr-PCR technique (Figure 1). Of the MRSA strains, 67% were classified into agr1 and 33% into agr3. Among the CL-MSSA strains, Of the CL-MSSA strains23 (42%) belonged to agr1, 14 (25%) to agr2, 11 (20%) to agr3, and 7 (13%) to agr4. Also, 9 (41%) of the C-MSSA strains belonged to agr1, followed by agr2 (8; 36%), agr3 (4; 18%) and agr4 (1; 5%) (see Figure 2).
Staphylococcal superantigen (SAg) gene profile among the isolates
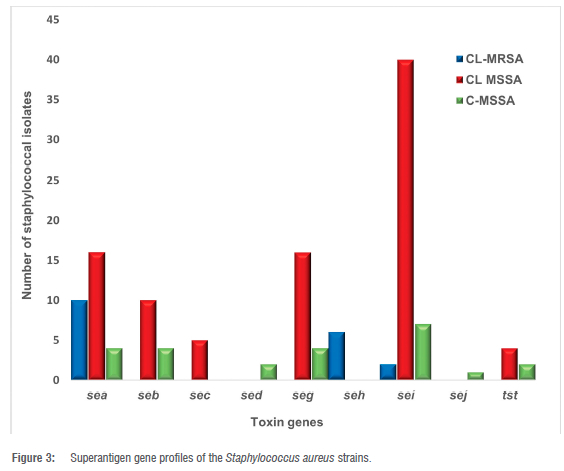
Eight (8) distinct SE genes (sea, seb, sec, sed, seg, seh, sei and sej) were recognised and none of the isolates was positive for see gene. Unlike the MSSA strains, only three types of SAg gene were detected among the MRSA strains, with sea accounting for the majority (10, 83%), followed by seh (6, 50%), and sei (2, 17%) (Figure 3). The 70 MSSA strains had sei (47, 67.1%) as the most prominent enterotoxin gene and sed was detected only in C-MSSA. CL-MSSA had 40 (60%) sei, 16 (22.9%) sea, 10 (14.3%) seb, 5 (7.1%) sec, 16 (22.9%) seg and no sed, seh nor sej were detected. In addition, 4 (5.7%) sea, seb, seg, 7 (10%) sei, 2 (2.9%) sed and 1 (1.4%) sej were identified amongst the C-MSSA strains. In all, 8 C-MSSA strains had no detectable SAg genes and the 6 tst genes discovered were peculiar to MSSA strains.
Enterotoxin genes co-existence and agr groups
In all, 18 CL-MSSA strains had 2 or 3 SAg gene combinations, with the highest number (sea-sei) occurring in 7 strains. Other combinations of the enterotoxin genes were also detected among the community strains; 5 MRSA strains co-harboured sea and seh genes and sea, seh and sei co-existed in 1 MRSA strain. The distribution of the toxin genes within the agr groups is shown in Table 2.
The sea and seh genes were associated with agr1 among the MRSA while sea, seh and sei were associated with agr3. For the CL-MSSA, sea, seb, sec, seg, seh and sei genes were connected to agr3, whereas sea, seb, sec, seg, sei and tst genes were related to agr4. Regarding C-MSSA, the sea, seb, sed and sei genes were associated with agr1, while sea and sei genes were found within agr2. seb, sed, seg, sei and tst genes were in agr3 and sea, seg and tst genes were associated with agr4. Two tst genes were found in agr1 and agr4 groups in the CL-MSSA, and one each in the agr3 and agr4 groups among the C-MSSA.
Distribution of agr genes and SAg in relation to specimen types
Whilst some specimens fitted into certain agr classes, all specimen types were represented in agr1 group except the ear/eye samples (Table 3). The most prevalent genes (sea and sei) were predominantly detected in pus (35%) and wound (50%) isolates. Table 4 shows other observed frequencies.
Discussion
In this study, the occurrence of methicillin resistance among the S. aureus isolates examined was 13%. Individuals harbouring MRSA, which is frequently multidrug resistant, are at risk of serious threat to themselves, and also play a role in the dissemination of the pathogen in hospital and community settings. Although the 29/307 MRSA recorded among hospitalised oncology patients examined in Ahvaz, Khuzestan Province, southwest of Iran22 was much lower, it could not be ruled out that the higher rate observed in this study is unconnected to the antibiotic use pattern in Nigeria.23 The isolates we examined were mostly from clinical sources and about 24% of the whole collection were from individuals who had no known indications for antibiotic use. The National Orthopaedic Hospital in Lagos has the capacity to accommodate up to 450 trauma patients, while the Lagos University Teaching Hospital, a leading tertiary hospital with over 761 admission beds, is the facility of last resort and referral for all disease conditions. As tertiary healthcare institutions, both hospitals may have a high prescription culture for antibiotics. Furthermore, antibiotics are widely accessible and can be purchased without a prescription over the counter. This has conferred selective pressure on the majority of bacterial pathogens in our environment.
With the exception of three non-typeable S. aureus isolates, all isolates examined in this study were defined into the four agr groups. In S. aureus, the accessory gene regulator (agr) plays a vital role in the temporal expression of a wide range of bacterial virulence factors. A large proportion of the MRSA strains (58.6%) fitted into the agr1 and agr3 clusters. The CL-MSSA and NS-MSSA strains, on the other hand, belonged to agr14. These outcomes are in contrast to those of a study conducted in Poland24 in which no agr4 was detected, but are comparable to the findings of Elazhari et al.21 Likewise, the majority (42%) of the isolates belonging to agr group 1 were MRSA, which was in agreement with the findings of other studies.20,23
In the present communication, the number of superantigen (SAg) genes detected varied significantly across the population of the S. aureus analysed, but was more pronounced among the methicillin-susceptible strains than their methicillin-resistant counterparts. The clinical relevance of this observation is that MSSA acquisition can be more deleterious. Elsewhere, Elazhari and others21 indicated that 19 of the MSSA strains examined in their study harboured SAg genes varying from 1 to 11, which supports our assumption. From another perspective, Ayeni et al.16 discovered neither seb or sec among their collection of S. aureus strains in southern Nigeria, which contradicts our findings. Additionally, our findings revealed a significant relationship between sea, seh and MRSA: 83% and 50% of MRSA strains had the sea and seh genes, respectively. In contrast, Ali et al.14 discovered the presence of the seb gene in MRSA strains. This signifies that the enterotoxin gene profiles of S. aureus may vary substantially depending on geographical area and population structure.25
We also found the simultaneous presence of sea and seh genes in five MRSA strains, as well as the occurrence of two or more distinct enterotoxin genes in 18 CL-MSSA. Four toxin genes (seb, seg, sei, tst) co-existed in one C-MSSA strain. The tst gene was exclusively present in CL-MSSA and C-MSSA isolates and was not detected in MRSA. Previously, Danelli and colleagues26 explained that tst carriage is commonly related with MSSA, which is consistent with our findings. Besides the possibility that MSSA isolates may have a lower genetic fitness burden because they do not have the SCCmec element27, Varshney et al.28 postulated that MSSA strains have an increased potential to secrete toxins than MRSA. S. aureus is known to have an outstanding array of virulence characteristics for initiating infections. This event may have major implications for public health.29 It has also been insinuated that even modest quantities of staphylococcal enterotoxins could elicit T-cell activation30, resulting in systemic infections including staphylococcal enterotoxin-induced shock and autoimmunity31. Similarly, Schmidt et al.32 speculated that TSST-1 expression was independent of S. aureus methicillin sensitivity. This could further explain the carriage of tst genes among the MSSA analysed in this study, albeit at low prevalence. Therefore, it is possible that there may be additional, as yet unexplained, variables that affect tst carriage. Further studies are needed to better understand TSST-1 expression mechanisms in S. aureus.
In our analysis, the agr1 group was represented in all specimens investigated, except for ear/eye samples, demonstrating some similarity with the results of Javdan and co-workers33. However, its dominance was higher in pus/wound and genitourinary tract samples. This is in contrast to the findings of Elazhari et al.21 These authors observed that agr2 and agr3 were prevalent in isolates from pus/wounds. This disparity between our findings and those of others may not be unconnected to sampling variability and study timeframe. While the agr'-MRSA had more enterotoxin genes, most of the toxigenic CL-MSSA and NS-MSSA belonged to agr3. The findings of this study also indicate that the expression of SAg genes was not unconnected to the types of specimens. The capacity to categorise infections based on sites or specimen types may give insight into the extent to which microorganisms play a role in disease initiation and progression. sei, the most prominent enterotoxin gene, was identified in all samples anaysed, but no seb, sec, seg, and seh were found among sputa isolates.
Meanwhile, the sej gene was only detected in sputum samples, while the tst genes were mainly present in pus/wound and nasal samples. Evidence from our study also shows that isolates that tested positive for sei and sea were significantly related with pus/wounds (p<0.05). This could alter the pathophysiology of wounds and present survival of the pathogen. Gergova et al.34 noted that more virulence genes were found in invasive S. aureus compared to isolates from non-invasive sites (nasopharyngeal secretion, skin lesion, urogenital tract and eye secretion). Their result corroborates our findings. They also reported that several of the genes (sei, sea, and seg) were found in individuals who had died from staphylococcal bacteraemia, underlining the possible serious clinical consequences of superantigenic S. aureus strains.
Conclusions
Our findings indicate that agr1 expression seems to be important for colonisation and establishment of S. aureus infections. Although the superantigen gene content of the pathogen differs significantly between MRSA and MSSA, the presence of the genes in MRSA poses an increased public health risk. The preponderance of MSSA and the connection of superantigenic genes may result in an expansion of strains with higher pathogenicity, which could lead to therapeutic dead ends. Future research may be required to determine the relationships between MSSA and tst carriage, as well as whether these associations are restricted to certain agr groups.
Acknowledgements
We are grateful to the staff of the Department of Microbiology, Otto-von-Guericke University, Germany, for technical support. We gratefully recognise Dr Beniam Ghebremedhin's invaluable professional support during the planning and development of the research. Financial aid was granted to N.V.E. by TETFUND (Tertiary Education Trust Fund, Grant No. ETF/ES/AST & D/UNIV/LAGOS/VOL.2, 2010).
Competing interests
We have no competing interests to declare.
Authors' contributions
N.V.E., S.A.A. and C.A.E.: Conceptualisation, methodology, data collection and analysis, writing - initial draft and proofreading. N.V.E., C.A.E., S.A.A., U.E.M.: Validation, project leadership, supervision. All authors read and approved the final manuscript.
References
1. Karsten B, Michael ZD, Robert LS. Staphylococcus, Micrococcus, and other catalase-positive cocci. In: Manual of clinical microbiology. 12th ed. Washington DC: ASM Press; 2021. p. 11-13. [ Links ]
2. Jakub MK, Alexander RH. Staphylococcus aureus bloodstream infections: Pathogenesis and regulatory mechanisms. Curr Opin Microbiol. 2020;53:51-60. https://doi.org/10.1016/j.mib.2020.02.005 [ Links ]
3. Mandeal SM, Ghosh AK, Pati BR. Dissemination of antibiotic resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant S. aureus strains isolated from hospital effluents. Am J Infect Control. 2015;43:87-88. https://doi.org/10.1016/j.ajic.2015.08.015 [ Links ]
4. Thammavongsa V Kim HK, Missiakos D, Schneewind O. Staphylococcal manipulation of host immune response. Nat Rev Microbiol. 2015;13(9):529-543. https://doi.org/10.1038/nrmicro3521 [ Links ]
5. Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: Molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23(2):214-224. https://doi.org/10.1016/j.chembiol.2016.01.004 [ Links ]
6. Jhalka K, Tara CS, Dipendra T. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. BioMed Res Int. 2014;2014:1-9. https://doi.org/10.1155/2014/827965 [ Links ]
7. Maleki DT, Ghalavand Z, Laabei M, Nikmanesh B, Houri H, Kodori M, et al. Molecular analysis of accessory gene regulator functionality and virulence genes in Staphylococcus aureus derived from paediatric wound infections. Infect Genet Evol. 2019;73:255-260. https://doi.org/10.1016/j.meegid.2019.05.013 [ Links ]
8. Tan L, Huang Y Shang W, Yang Y Peng H, Hu Z, et al. Accessory gene regulator (agr) allelic variants in cognate Staphylococcus aureus strain display similar phenotypes. Front Microbiol. 2022;13:700894. https://doi.org/10.3389/fmicb.2022.700894 [ Links ]
9. Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631-641. https://doi.org/10.1128/IAI.70.2.631-641.2002 [ Links ]
10. Singh R, Ray P Quorum sensing-mediated of staphylococcal virulence and antibiotic resistance. Future Microbiol. 2014;9:669-681. https://doi.org/10.2217/fmb.14.31 [ Links ]
11. Tahmasebi H, Dehbashi S, Arabestani MR. Association between the accessory gene regulator (agr) locus and the presence of superantigen genes in clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res Notes. 2019;12:130. https://doi.org/10.1186/s13104-019-4166-7 [ Links ]
12. Christian J, Alexander RH. Regulation of Staphylococcus aureus virulence. Microbiol Spectr. 2019;7(2):10. https://doi.org/10.1128/microbiolspec.GPP3-0031-2018 [ Links ]
13. Gordon YCC, Justin SB, Michael O. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547-569. https://doi.org/10.1080/21505594.2021.1878688 [ Links ]
14. Yin X, Su-Yun Q, Kai-Hu Y Fang D, Wen-Qi S, Chen S, et al. Clinical and molecular characteristics of Staphylococcus aureus isolated from Chinese children: Association among the agr groups and genotypes, virulence genes and disease types. World J Pediatr. 2021;17:180-188. https://doi.org/10.1007/s12519-021-00421-4 [ Links ]
15. Choopani A, Heiat M, Amini E, Golpuch M, Aghamollaei H. The relationship between the presence of enterotoxin type b gene and antibiotic resistance in Staphylococcus aureus. J Appl Biotechnol Rep. 2015;2(1):203-206. [ Links ]
16. Ayeni FA, Ruppitsch W, Allerberger F. Molecular characterization of clonal lineage and staphylococcal toxin genes from S. aureus in Southern Nigeria. Peer J. 2018;6, e5204. https://doi.org/10.7717/peerj.5204 [ Links ]
17. Akinduti AP Osiyemi JA, Banjo TT, Ejilude O, El-Ashker M, Adeyemi AG, et al. Clonal diversity and spatial dissemination of multi-antibiotics resistant Staphylococcus aureus pathotypes in Southwest Nigeria. PLoS ONE. 2021;16(2), e0247013. https://doi.org/10.1371/journal.pone.0247013 [ Links ]
18. Enwuru NV Adesida SA, Enwuru CA, Ghebremedhin B, Mendie UE, Coker AO. Genetics of bi-component leukocidin and drug resistance in nasal and clinical Staphylococcus aureus in Lagos, Nigeria. Microb Pathog. 2018;115:1-7. https://doi.org/10.1016/j.micpath.2017.12.030 [ Links ]
19. Ender M, Berger-Bachi B, McCallumn N. Variability in SCCmecN1 spreading among injection drug users in Zurich, Switzerland. BMC Microbiol. 2007;7:62. https://doi.org/10.1186/1471-2180-7-62 [ Links ]
20. Shopsin B, Mathema B, Alcabes P Said-Salim B, Lina G, Matsuka A, et al. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J Clin Microbiol. 2003;41(1):456-459. https://doi.org/10.1128/JCM.41.L456-459.2003 [ Links ]
21. Elazhari M, Elhabchi D, Zerouali K, Dersi N, Elmalki A, Hassar M, et al. Prevalence and distribution of superantigen toxin genes in clinical community isolates of Staphylococcus aureus. J Bacteriol Parasitol. 2011;2:107. https://doi.org/10.4172/2155-9597.1000107 [ Links ]
22. Montazeri EA, Khosravi AD, Khazaei S, Sabbagh A. Prevalence of methicillin resistance and superantigenic toxins in Staphylococcus aureus strains isolated from patients with cancer. BMC Microbiol. 2021;21:262. https://doi.org/10.1186/s12866-021-02319-7 [ Links ]
23. Chukwu EE, Oladele DA, Enwuru CA, Gogwan PL, Abuh D, Audu RA, et al. Antimicrobial resistance awareness and antibiotic prescribing behavior among healthcare workers in Nigeria: A national survey. BMC Infect Dis. 2021;21(1):22. https://doi.org/10.1186/s12879-020-05689-x [ Links ]
24. Grazul M, Balcerczak E, Sienkiewicz M. Analysis of the presence of the virulence and regulation genes from Staphylococcus aureus (S. aureus) in coagulase negative staphylococci and the influence of the staphylococcal cross-talk on their functions. Int J Environ Res Public Health. 2023;20(6):5155. https://doi.org/10.3390/ijerph20065155 [ Links ]
25. Kolawole DO, Adeyanju A, Schaumburg F, Akinyoola AL, Lawal OO, Amusa YB, et al. Characterization of colonizing Staphylococcus aureus isolated from surgical wards' patients in a Nigerian university hospital. PLoS ONE. 2013;8(7), e68721. https://doi.org/10.1371/journal.pone.0068721 [ Links ]
26. Danelli T, Duarte FC, de Oliveira TA, da Silva RS, Alfieri DF, Gonçalves GB, et al. Nasal carriage by Staphylococcus aureus among healthcare workers and students attending a university hospital in southern Brazil: prevalence, phenotypic, and molecular characteristics. Interdiscip Perspect Infect Dis. 2020;2020:3808036. https://doi.org/10.1155/2020/3808036 [ Links ]
27. Zhao H, Xu S, Yang H, He C, Xu X, Hu F, et al. Molecular typing and variations in amount of tst gene expression of TSST-1-producing clinical Staphylococcus aureus isolates. Front Microbiol. 2019;10(1388). https://doi.org/10.3389/fmicb.2019.01388 [ Links ]
28. Varshney AK, Martinez LR, Hamilton SM, Bryant AE, Levi MH, Gialanella P, et al. Augmented production of Panton-Valentine leukocidin toxin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus is associated with worse outcome in a murine skin infection model. J Infect Dis. 2010;201:92-96. https://doi.org/10.1086/648613 [ Links ]
29. Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547-569. https://doi.org/10.1080/21505594.2021.1878688 [ Links ]
30. Pinchuk IV Beswick EJ, Reyes VE. Staphylococcal enterotoxins. Toxins. 2010;2(8):2177-2197. https://doi.org/10.3390/toxins2082177 [ Links ]
31. Principato M, Qian BF. Staphylococcal enterotoxins in the etiopathogenesis of mucosal autoimmunity within the gastrointestinal tract. Toxins. 2014;6(5):1471-1489. https://doi.org/10.3390/toxins6051471 [ Links ]
32. Schmidt KA, Manna AC, Gill S, Cheung AL. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun. 2001;69:4749-4758. https://doi.org/10.1128/IAI.69.8.4749-4758.2001 [ Links ]
33. Javdan S, Narimani T, Abadi MSS, Gholipour A. Agr typing of Staphylococcus aureus species isolated from clinical samples in training hospitals of Isfahan and Shahrekord. BMC Res Notes. 2019;12:363. https://doi.org/10.1186/s13104-019-4396-8 [ Links ]
34. Gergova RT, Tsitou VS, Gergova II, Muhtarova AA, Mitov IG. Correlation of methicillin resistance and virulence genes of Staphylococcus aureus with infection types and mode of acquisition in Sofia, Bulgaria. Afr J Clin Exper Microbiol. 2019;20(4):280-288. https://doi.org/10.4314/ajcem.v20i4.3 [ Links ]
 Correspondence:
Correspondence:
Solayide Adesida
Email: sadesida@unilag.edu.ng
Received: 23 Apr. 2021
Revised: 05 July 2023
Accepted: 08 July 2023
Published: 28 Sep. 2023
Editor: Pascal Bessong
Funding: Tertiary Education Trust Fund (ETF/ES/AST, D/UNIV/LAGOS/VOL.2, 2010)














