Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.119 n.5-6 Pretoria May./Jun. 2023
http://dx.doi.org/10.17159/sajs.2023/14659
PERSPECTIVE
Managing and assembling population-scale data streams, tools and workflows to plan for future pandemics within the INFORM-Africa Consortium
Jenicca PoongavananI; Joicymara XavierII, III, IV; Marcel DunaiskiV; Houriiyah TegallyI, VI; Sunday O. OladejoI; Olawole AyorindeVII; Eduan WilkinsonI; Cheryl BaxterI, VIII; Tulio de OliveiraI, VI, VIII, IX
ICentre for Epidemic Response and Innovation (CeRI), School for Data Science and Computational Thinking, Stellenbosch University, Stellenbosch, South Africa
IIInstitute of Agricultural Sciences, Federal University of Vales do Jequitinhonha e Mucuri, Unaí, Brazil
IIIRené Rachou Institute, Oswaldo Cruz Foundation, Belo Horizonte, Brazil
IVInstitute of Biological Science, Federal University of Minas Gerais, Belo Horizonte, Brazil
VDepartment of Computer Science, School for Data Science and Computational Thinking, Stellenbosch University, Stellenbosch, South Africa
VIKwaZulu-Natal Research Innovation and Sequencing Platform (KRISP), Nelson R Mandela School of Medicine, University of KwaZulu-Natal, Durban, South Africa
VIIInstitute of Human Virology, Abuja, Nigeria
VIIICentre for the AIDS Programme of Research in South Africa (CAPRISA), Durban, South Africa
IXDepartment of Global Health, University of Washington, Seattle, WA, USA
ABSTRACT
SIGNIFICANCE:
The INFORM-Africa Consortium, a research hub of the NIH-funded DS-I Africa, will leverage the Data Management and Analysis Core (DMAC) and Next Generation Sequencing (NGS) Core to ensure effective data management and analysis. The DMAC will capture and analyse data, making it accessible to collaborators across multiple African countries and future research hubs. The aim is to increase access to high-quality, reproducible data that can be used to engage policymakers and better prepare for future pandemics, while also removing barriers to data sharing and integration across institutions. Ultimately, this goal will facilitate data-driven decision-making and advance public health initiatives.
Keywords: INFORM-Africa data management, collaboration, SARS-CoV-2, HIV
Introduction
The SARS-CoV-2 virus has caused over 12 million recorded cases of COVID-19 in Africa with over 256 000 lives claimed.1 The rapid growth of COVID-19 to pandemic proportions in Africa occurred against a backdrop of existing epidemics of HIV, tuberculosis and malaria and a rising burden of non-communicable diseases, which placed additional demands on already strained healthcare systems. In 2020, an initial survey by the World Health Organization (WHO), using clinical and epidemiological data predominantly from South Africa, suggested that people living with HIV were 30% more likely to die from COVID-19 among those hospitalised with the disease.2,3 On the other hand, some reports indicate that HIV infection itself does not appear to be a risk factor for severe COVID-19.4,5 Individually and collectively, these studies do not provide sufficient data, due largely to their limited sample sizes, to understand the relationship between SARS-CoV-2 infection and HIV. In order to create a core capacity for governments across Africa to better respond to current and future epidemics, it is crucial to understand the synergies at work between the two diseases at a population scale.
To better address issues around public health, it is important to develop the capacity to effectively generate, collect, store, clean, annotate, link, and share data from diverse sources. Furthermore, a research gap in epidemic modelling around the world, and specifically in Africa, is the lack of population-scale epidemiologic data sources, properly annotated and linked across health services. Between continent-wide technical and infrastructural resource limitations and fragile health systems, the need for population-scale epidemiological and frequently updated data, is even more urgent to inform interventions rapidly.6,7 Consequently, 'The Role of Data Streams in Informing infection Dynamics in Africa' (INFORM-Africa) Research Hub was established; this Hub focuses on the effective use of big data from South Africa and Nigeria as a cornerstone of future pandemic preparedness.
The INFORM-Africa Hub consists of three main project groups: Project 1 focuses on how viral genomic variation, adoption of public health mitigation measures, and mobility patterns contribute to spatially and temporally explicit pathways of SARS-CoV-2 transmission at local and regional scales. Project 2 examines the effect of movement-based restrictions on mobility in Nigeria and South Africa, compares pre-pandemic to post-pandemic movement patterns using cell phone mobility data, and associates specific movement patterns with COVID-19 risk factors. This model incorporates state-of-the-art mobility analytics from the transportation sector, applying them to the African context, possibly for the first time. Project 3 studies the interplay between SARS-CoV-2 and HIV in the two countries most impacted by the syndemics in Africa. It investigates to what extent shared geospatial, mobility and demographic factors affect risk of both infections and how each infection affects the outcomes of the other, and whether the host genetic variation in Africa explains the COVID-19 outcomes in Africa.
During the COVID-19 pandemic, public health data has been employed to gain insight, track, and limit the spread of the virus. There are several institutions that have been collecting, managing and analysing clinical and epidemiological data in Africa, such as South Africa's National Institute for Communicable Diseases (NiCD), which is a division of the National Health Laboratory Service (NHLS) in South Africa that conducts surveillance, outbreak investigations and research on communicable diseases. They collect and analyse epidemiological data to monitor the incidence and prevalence of diseases and to guide public health intervention. There is also the South African Medical Research Council (SAMRC) which has a wealth of information. They conduct research on a wide range of health issues, including infectious diseases, non-communicable diseases, and injury-related deaths. They also annually publish a report called the 'South African Burden of Disease Study' which provides detailed information on death and disability in South Africa. One of the SAMRC's key contributions during the pandemic was providing regular updates on the country's COVID-19 status. Stats-SA have also been collating data by conducting household surveys and collecting healthcare data, and at the same time providing statistical analysis.
Moreover, in order for policy decisions to be effective, we need to consider a holistic understanding of the epidemiological situation, including scientific data, as well as the broader context in which the disease is spreading. This context might include factors such as the availability of healthcare resources, the socio-economic impact of public health measures, and the political will to implement effective policies. In addition, the question of improving the reliability and quality of epidemiological data for morbidity, mortality and sero-prevalence in community and hospital settings as well as for understanding the impact of preventive measures, such as vaccination and other measures through timely and targeted representative sampling methods, becomes crucial. While mobility and infectious agent genomics cannot influence policy alone, they are key factors that need to be integrated into a robust epidemiological data landscape to obtain broader understanding of transmission dynamics and inform effective policy decisions that can help control the spread of infectious diseases.
It is evident that data availability primes research and discovery in the sciences, but the global pandemic coverage has also propelled the engagement with public health data, and data in general, into the public discourse. Data ranging from genomic, patient management and mobility data are crucial for the respective projects to answer the questions they are investigating. However, key challenges include obtaining relevant genomic data and metadata together with patient data, integrating these data originating from multiple sources, applying efficient computational algorithms to cope with these large data sets, and establishing sampling frameworks to enable robust conclusions.
Data sharing amongst data custodians can be contentious and often involves navigating complex policy restrictions and political dynamics. The 2020 State of Open Data report identified trust (or the lack thereof) as a key barrier to data sharing.8 To help the INFORM-Africa Research Hub navigate through the ocean of multiple data streams, a Data Management and Analysis Core (DMAC) and Next Generation Sequencing (NGS) core was established. DMAC's responsibility is to address issues of trust, together with managing institutional policies on ethics, intellectual property rights and data ownership agreements -a challenge that requires innovative approaches on data access policies.
The DMAC and NGS Core
The DMAC and NGS core play a key role in assembling and managing the INFORM-Africa Research Hub's data and in providing seamless access to a set of tools and workflows as well as generating next generation sequencing data. The DMAC intends to empower the INFORM-Africa Research Hub by expanding data science research opportunities and capacity in Africa through the involvement of early-stage investigators and trainees, and the data science training and support provided within the INFORM-Africa and across the DS-I Africa Consortium.
The DMAC leverages state-of-the-art computing platforms and uses integrative data analysis frameworks to support the INFORM-Africa Research Hub. The core will accommodate multiple data types (ranging from existing population-scale individual-level clinical data and genomic data to geospatial and mobility data) and additional resources, such as standard operating procedures, protocols and training materials9 based on the FAIR principles for scientific data management and stewardship to improve the Findability, Accessibility, Interoperability and Reuse (reproducibility) of data. Guided by these principles, the overarching goal is to provide the Research Hub with a unified environment for data management, computation, and technical support for collaborative work within and between projects in the INFORM-Africa Hub.
To date, our researchers have been using the traditional model of data analysis in which they are required to download their data from centralised data warehouses onto their local computers, install and maintain their suite of computational tools, and execute analyses using local computing resources. Following the traditional model, each project within the consortium would have to establish and maintain its own data centres, which would create major administrative inefficiencies such as the duplication of data and analysis tools that must be deployed and maintained separately within each centre. The data management tasks also become unsustainable given the unprecedented quantity of genomic and epidemiological data and the frequency at which these data are updated. Furthermore, many data analysis tasks using cutting-edge computational models are impracticable due to the scale of their data requirements and computational complexities when relying on the traditional computation paradigm.
The DMAC will progressively shift towards a more contemporary approach by moving to the cloud. Cloud computing as defined by the US National Institute of Standards and Technology is:
...a model for enabling convenient, on-demand network access to a shared pool of configurable computing resources (e.g. networks, servers, storage, applications, and services) that can be rapidly provisioned and released with minimal management effort or service provider interaction.10
This definition goes hand in hand with the DMAC's goals and vision within the INFORM-Africa Research Hub.
With the massive volumes of data that we are expecting, integrating multiple genomic, epidemiological and patient data sources can be a complex process and requires techniques to resolve inconsistencies in temporal structure and encoding. To overcome these challenges, we have established a data lake architecture to guide an effective curation process. Data lakes have recently emerged as an enterprise solution to manage large amounts of heterogeneous data for modern data analytics. A data lake architecture can be described as a schema-free repository that allows users to store structured, unstructured, and unprocessed data at any scale, based on cloud computing.5,11,12 The main advantage of choosing a data lake architecture as a data management paradigm is the increased flexibility in terms of data type support, as well as the ability to more easily cater to the specific data needs of various users. This will allow the DMAC to continuously support and easily adapt to the requirements of the diverse projects that will be hosted on the DMAC and NGS platform. The DMAC's integrative data management and analysis strategy is summarised in Figure 1.
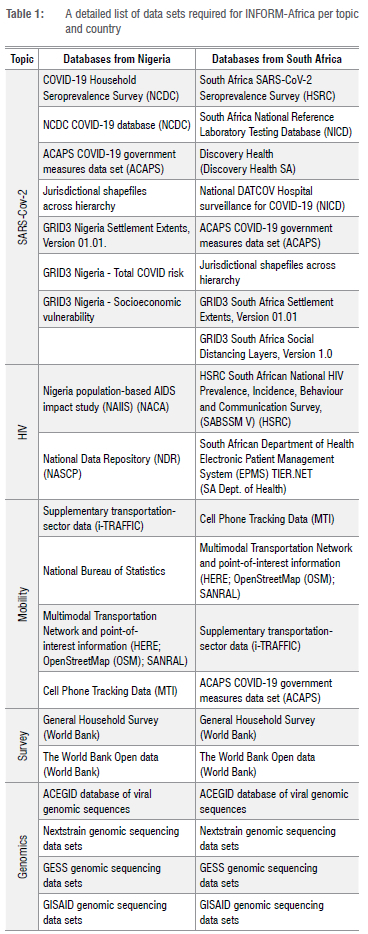
The data panel in Figure 1 shows the diverse data types that the Hub necessitates and the different data sources that the DMAC and NGS core are responsible for integrating. All the data sources are described in detail in Table 1. The architecture will allow for efficient ingestion of any data types such as genomic files, epidemiological data, or GPS data, while supporting several data access types such as streamed data, batch file uploads, or API access. These are very different data types, each of which requires different standards, formats and storage.
In terms of data governance, access to data is made possible through signed data sharing agreements with public and private providers. We also work closely with the data providers to establish an efficient path for data transfers and updates to existing data sets. Several data sets have already been assembled from various data warehouse sources across Africa. The NGS core contributes toward generating genomic data and metadata that the Research Hub would require.
Once the raw data are acquired, they are deposited on a distributed file system before being curated by the DMAC team. All sensitive data are anonymised and, where necessary, encrypted for storage and transfer. The curation process encompasses extracting and transforming the data into a format that each project within the INFORM-Africa group can use to run their respective analyses. Data curation and quality control measures occur regularly, following a standard protocol for data monitoring and addressing any identified issues, by involving data providers and project investigators.
Once transformed and validated, the data are stored and automatically shared on a data platform built in the cloud that allows extensive collaborative genomic research. The workspace will provide well-established tools to easily filter the data, perform and share analyses. By using a cloud-based data management platform, the DMAC's aim is to create workspaces dedicated to the iNFORM-Africa consortium. Through these workspaces, we are able to enforce user access control protocols as required by the various projects.
The DMAC is also invested in sharing high-quality tools and workflows for use in the Research Hub. Dockstore (https://dockstore.org/) is a workflow and tool publishing platform that is widely used in the bioinformatics and genomics community. Dockstores can be leveraged both as a source of high-quality workflows and tools as well as a distribution platform for tools and workflows produced by the DMAC team. The goal is to share these workflows with researchers across the Hub so that they can easily use these workspaces.
The DMAC will provide training and support to help researchers develop the platform in line with their needs. The training and support will encompass a variety of topics including data quality assurance and quality control training, especially for early-stage investigators and trainees in the iNFORM-Africa Hub. The DMAC and NGS core will also provide support for all new data collection, to ensure uniform data entry procedures and data quality across Hub partners. Additionally, after performing a needs analysis, we will implement an agile data science training programme that includes big data and bioinformatics analysis to train a broad range of stakeholders to manage, process, analyse and interpret biological data together with geospatial and other relevant data as needed.
Conclusion
in summary, the DMAC and NGS core are an essential link between the projects of the iNFORM-Africa Research Hub. The DMAC and NGS core will facilitate the seamless integration and linking of various public and private data sets, access to new data and tools created by the projects and cores, and broader sharing, including with the Research Hub and the DS-I Africa Consortium. By using a cloud-based platform, our focus is to enable high-level and reproducible data analysis and cross-network projects between collaborators across the three projects to achieve the overall goal of the INFORM-Africa Research Hub. The platform will enable biological discovery from the big data that is available in Africa. Finally, the DMAC and NGS core will contribute toward the INFORM-Africa aims by expanding data science research opportunities and building capacity throughout Africa.
Acknowledgements
We acknowledge support from the US National Institutes of Health through the INFORM-Africa project that is administered by IHVN (U54 TW012041) and the eLwazi Open Data Science Platform and Coordinating Center (U2CEB032224).
Competing interests
We have no competing interests to declare.
References
1. Ritchie H, Mathieu E, Rodés-Guirao L, Appel C, Giattino C, Ortiz-Ospina E, et al. Our world in data: Coronavirus pandemic (COVID-19) [webpage on the Internet]. c2020 [cited 2022 Aug 10]. Available from: https://ourworldindata.org/coronavirus [ Links ]
2. Msomi N, Lessells R, Mlisana K, de Oliveira T. Africa: Tackle HIV and COVID-19 together. Nature. 2021;600(7887):33-36. https://doi.org/10.1038/d41586-021-03546-8 [ Links ]
3. World Health Organization (WHO). Clinical features and prognostic factors of COVID-19 in people living with HIV hospitalized with suspected or confirmed SARS-CoV-2 infection, 15 July 2021 [data set]. https://apps.who.int/iris/handle/10665/342697 [ Links ]
4. Brown LB, Spinelli MA, Gandhi M. The interplay between HIV and COVID-19: Summary of the data and responses to date. Curr Opin HIV AIDS. 2021;16(1):63-73. https://doi.org/10.1097/COH.0000000000000659 [ Links ]
5. Eisinger RW, Lerner AM, Fauci AS. Human immunodeficiency virus/AIDs in the era of coronavirus disease 2019: A juxtaposition of 2 pandemics. J Infect Dis. 2021;224(9):1455-1461. https://doi.org/10.1093/infdis/jiab114 [ Links ]
6. Kucharski AJ, Hodcroft EB, Kraemer MUG. Sharing, synthesis and sustainability of data analysis for epidemic preparedness in Europe. Lancet Reg Health Eur. 2021;9, Art. #100215. https://doi.org/10.1016/jJanepe.2021.100215 [ Links ]
7. Khan MS, Dar O, Erondu NA, Rahman-Shepherd A, Hollmann L, Ihekweazu C, et al. Using critical information to strengthen pandemic preparedness: The role of national public health agencies. BMJ Glob Health. 2020;5(9), e002830. https://doi.org/10.1136/bmjgh-2020-002830 [ Links ]
8. Porter SJ, Hook DW. How COVID-19 is changing research culture. London: Digital Science; 2020. [ Links ]
9. Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3, Art. #160018. [ Links ]
10. Mell P, Grance T. The NIST definition of cloud computing. Gaithersburg, MD: National Institute of Standards and Technology; 2011. https://doi.org/10.6028/NIST.SP.800-145 [ Links ]
11. Giebler C, Gröger C, Hoos E, Schwarz H, Mitschang B. Leveraging the data lake: Current state and challenges. In: Ordonez C, Song I-Y Anderst-Kotsis G, Tjoa AM, Khalil I, editors. Big data analytics and knowledge discovery: 21st International Conference, DaWaK 2019; 2019 August 26-29; Linz, Austria. Cham: Springer International Publishing; 2019. p. 179-188. https://doi.org/10.1007/978-3-030-27520-4_13 [ Links ]
12. Ordonez C, Song I-Y Anderst-Kotsis G, Tjoa AM, Khalil I, editors. Big data analytics and knowledge discovery: 21st International Conference, DaWaK 2019; 2019 August 26-29; Linz, Austria. Cham: Springer International Publishing; 2019. https://doi.org/10.1007/978-3-030-27520-4 [ Links ]
 Correspondence:
Correspondence:
Jenicca Poongavanan
Email: jenicca1193@gmail.com
Published: 30 May 2023
Funding: US National Institutes of Health through INFORM-Africa (U54 TW012041), eLwazi Open Data Science Platform and Coordinating Center (U2CEB032224)














