Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.119 no.3-4 Pretoria Mar./Abr. 2023
http://dx.doi.org/10.17159/sajs.2023/12064
RESEARCH ARTICLE
Evaluating 3D human face reconstruction from a frontal 2D image, focusing on facial regions associated with foetal alcohol syndrome
Felix AtuhaireI; Bernhard EggerII, III; Tinashe MutsvangwaIV
IDepartment of Biomedical Sciences and Engineering. Mbarara University of Science and Technology, Mbarara. Uganda
IIDepartment of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge. Massachusetts, USA
IIIDepartment of Computer Science. Friedrich-Alexander-Universität Erlangen-Nürnberg. Erlangen Germany
IVDivision of Biomedical Engineering. Department of Human Biology, University of Cape Town, Cape Town. South Africa
ABSTRACT
Foetal alcohol syndrome (FAS) is a preventable condition caused by maternal alcohol consumption during pregnancy. The FAS facial phenotype is an important factor for diagnosis, alongside central nervous system impairments and growth abnormalities. Current methods for analysing the FAS facial phenotype rely on 3D facial image data, obtained from costly and complex surface scanning devices. An alternative is to use 2D images, which are easy to acquire with a digital camera or smart phone. However, 2D images lack the geometric accuracy required for accurate facial shape analysis. Our research offers a solution through the reconstruction of 3D human faces from single or multiple 2D images. We have developed a framework for evaluating 3D human face reconstruction from a single-input 2D image using a 3D face model for potential use in FAS assessment. We first built a generative morphable model of the face from a database of registered 3D face scans with diverse skin tones. Then we applied this model to reconstruct 3D face surfaces from single frontal images using a model-driven sampling algorithm. The accuracy of the predicted 3D face shapes was evaluated in terms of surface reconstruction error and the accuracy of FAS-relevant landmark locations and distances. Results show an average root mean square error of 2.62 mm. Our framework has the potential to estimate 3D landmark positions for parts of the face associated with the FAS facial phenotype. Future work aims to improve on the accuracy and adapt the approach for use in clinical settings.
SIGNIFICANCE: Our study presents a framework for constructing and evaluating a 3D face model from 2D face scans and evaluating the accuracy of 3D face shape predictions from single images. The results indicate low generalisation error and comparability to other studies. The reconstructions also provide insight into specific regions of the face relevant to FAS diagnosis. The proposed approach presents a potential cost-effective and easily accessible imaging tool for FAS screening, yet its clinical application needs further research
Introduction
Early detection of foetal alcohol syndrome (FAS) allows for early intervention, mitigates the onset of secondary disorders such as mental breakdown or improper sexual behaviours, and leads to significantly better clinical outcomes.1 The diagnosis of FAS is based on the evidence of central nervous system abnormalities, evidence of growth abnormalities, and a characteristic pattern of facial anomalies, specifically short palpebral fissure length, smooth philtrum, flat upper lip, and flat midface.2,3 The FAS facial phenotype has been emphasised clinically for diagnosis.4-7 However, clinical evaluation requires the expertise of trained dysmorphologists. This requirement limits efforts for large-scale screening in suspected high prevalence regions, such as South Africa, which has a prevalence rate estimated to be between 93 and 128 per 1000 live births8, and a shortage of highly trained clinical personnel. Alternative methods for assessing the FAS facial phenotype are possible but require careful acquisition of face data. Face data collection methods include direct anthropometry using handheld rulers and callipers. Indirect anthropometry, on the other hand, is possible through the acquisition of face data through 2D photogrammetry, 3D stereophotogrammetry, and 3D surface imaging scanners.2,9,10 Direct anthropometry introduces inaccuracies due to the indentation of some features during contact measurements with physical instruments. For this reason, more efforts have been put into indirect anthropometry, which has the added benefit of near-instantaneous patient data acquisition. Furthermore, with indirect approaches, measurements on the images can be repeated in the absence of subjects. Indirect evaluation on 3D image data is typically more accurate than on 2D images.11 However, acquiring 3D face images using 3D surface scanners tends to be costly and precludes large-scale deployment in low-resource settings.
Reconstruction of the 3D human face from a single 2D image is a popular topic of research, with applications in face recognition, face tracking, face animation, and medical analysis of faces.12 However, to date, there has not been any report on the quantitative suitability of 3D from 2D face reconstruction for FAS-related facial phenotype characterisation. In this study, our aim was to evaluate the geometric accuracy of a 3D human face reconstruction from a single 2D facial image, using a 3D morphable model of the face.13 We focused on 3D reconstruction of the complete face to enable surface-based approaches, and to allow us to evaluate landmark and distance-based measurements. We tested if such a reconstruction algorithm could be suitable for automated analysis of facial features related to FAS.
Related work
Three-dimensional morphable models (3DMMs) are high-resolution generative models containing shape and texture variations from sample populations.13-17 Typically, 3DMMs are built from a set of 3D face scans after establishing anatomical dense correspondences across the face data set. Establishing correspondences ensures that similar features across a set of 3D face scans match each other (e.g. the tip of the nose or the eye corners) - we call this process 'registration'.
Several methods for building 3DMMs from a set of 3D face scans have been presented over the years.12 In pioneering work, Blanz and Vetter13 built a 3DMM from a set of face scans after computing dense correspondences with an optical flow-based registration technique. The shape and texture variations in a collection of face scans were then modelled using principal component analysis (PCA), resulting in a low dimensional representation. The learned face models were used to estimate a 3D face surface from a single 2D face image. Early 3DMMs were built using just hundreds of face scans. However, a recent study by Booth et al.16 constructed a 3DMM known as the Large-Scale Facial Model using 9663 3D facial scans. Booth et al.16 used the non-rigid iterative closest point (NICP) algorithm18 for registration of the template face surface to each target face scan in the data set, aided by generalised Procrustes analysis (GPA) for similarity alignment of the registered face scans. They then used PCA19 for statistical analysis of the registered face scans. The 3DMMs have already been successfully applied in various application areas including face tracking, face recognition, face segmentation, and face reconstruction.12 However, additional research focusing on human face variations would still be required before the morphable model could be used for medical purposes.16
Blanz and Vetter's13 work was seminal, but their approximate 3D face meshes were only qualitatively evaluated. Romdhani and Vetter20 took a different approach, extracting multiple features from a single image. The extracted features were then used to estimate a 3D face surface by minimising a cost function. In 2009, 3DMM, the Basel face model, was made available for research purposes and enabled the community to grow faster.21 Aldrian and Smith22 developed the first publicly available inverse graphics algorithm based on a 3DMM. Schonborn et al.23 employed a sampling-based approach to fit a Gaussian process morphable model to a single 2D image. The face shape reconstruction accuracy as measured by a root mean squared average was 3.79 mm. Recently, a first benchmark was established for 3D reconstruction from 2D images.24 This benchmark is, however, strongly biased towards light skin tones, which is a narrow subset of the world's population and might not be representative for general clinical application. The state-of-the-art method on this benchmark is a deep learning based method for 3DMM reconstruction, with an average reconstruction error of 1.38 mm.25
While reconstruction algorithms are reported in the literature, there is limited research evaluating the accuracy of these algorithms, which has implications for the algorithm performance on medical-related applications. Additionally, to the best of our knowledge, model-based 3D face reconstruction from 2D image approach has not been evaluated with a focus on FAS applications, perhaps because 3D ground truth data may not be available. A robust single image-based reconstruction approach could offer a cost-effective alternative to 3D surface capturing systems.
Methods
Data description
We based our experiments on the BU-3DFE face database, which is a publicly available data set of high-quality 3D scans, acquired using the 3dMD face system.26 It consists of face scans of 98 subjects of different ethnicities (56 female and 42 male subjects aged between 18 and 70 years). We used only the facial scan with a neutral expression for each identity (see Figure 1 for an example of the images). The data were used with ethical approval from both the University of Cape Town and the State University of New York. To maximise the number of faces for training, we performed a leave-one-out cross-validation scheme for our experiments. From each face scan, we derived the 3D ground truth face shape, established correspondence to our model template, and rendered a frontal 2D image for our 2D to 3D reconstruction task. To reach maximal accuracy, we used 12 manual landmarks to initialise the 2D to 3D reconstruction process: right outer and inner canthi, glabella, left inner and outer canthi, right and left alares, pronasale, subnasale, right and left cheilions. We did not rely strictly on these landmarks as the fitting framework used has been shown to work with automatic landmark detection. This gave us a set of 2D images with known ground truth 3D shapes for learning and evaluating our model and reconstruction framework.

Rigid alignment of face scans
The goal of rigid alignment is to bring all the face scans into a common coordinate system without deformations. Given a set of pre-processed 3D face scans (pre-processing involves trimming the face scans to remove the unwanted regions such as the hair and neck regions) and a set of facial landmarks for each face scan, the facial landmarks were used to calculate a least-squares alignment that brought landmarks corresponding across scans as close together as possible (Procrustes alignment). The training face scans were mapped, using rigid transformations, to the mean of the Basel face model27, which represents a common reference face surface. The results of these alignments are a collection of rigidly aligned 3D face scans.
Registration of face scans
After rigid alignment, we used a deformable model to establish dense correspondences between a reference face surface and each target face scan in the training data. By dense correspondences, we mean finding the mappings between similar features across the data set. The goal of registration is to re-parameterise the face scans to have the same number of vertices and triangulations across face scans in the training set with the key feature that each vertex corresponds to the same point on each face. The reference face surface is fitted to each target face scan, using a Gaussian process fitting approach27, to obtain dense face surface deformations, which best match a target face scan to a common reference face surface. The time for registering the reference to each target face was 5-8 minutes computed on an Intel(R) Core (TM) Í5-8350U CPU @ 1.7 GHz. This registration approach builds on a Gaussian process defined by mean and covariance functions to model smooth deformations of the template shape.28 29 During registration, we searched the optimal set of parameters of our Gaussian process model to match the 3D scan at hand. The results of applying the fitting approach are registered 3D face scans. As we wanted to build a 3D MM, at this stage, we also extracted the colour per vertex from the closest vertex on the face scan - this enabled us to not only build a shape but also a texture model.
Building face models
With dense correspondences established among the training data, we removed translations and rotations on the data to retain shape deformation. To perform these non-shape-related transformations on the training data, we applied the Procrustes analysis approach.30-32 After removing alignments, the principal modes of variation were extracted from training data using PCA28,29 to build 3D morphable face models (3DMMs). The 3DMMs consisted of the face means and the principal components as modes of variation. The 3DMMs are expressed as linear combinations of shape and texture vectors in the face subspace. The time required to build a surface model from each registered face scan was 10-20 minutes computed on an Intel(R) Core (TM) I5-8350U CPU @ 1.7 GHz. An example of registration results as well as a resulting 3DMM built from all 98 scans are illustrated in Figure 2.
3D from 2D face reconstruction
The key application of a 3DMM that we were interested in was the estimation of 3D face surfaces and 3D landmark positions for FAS measurements from 2D images. In the reconstruction setting, the 3DMM acts as a prior of 3D face shape, and we searched for the most likely reconstruction given only 2D images. This is potentially useful because 2D images, in contrast to 3D scans, are easy to acquire using either a mobile phone or a portable camera. One of the goals of this study was to reconstruct a neutral 3D face with shape and texture information from a single frontal 2D image and evaluate how close that reconstruction was to the known ground truth.
Given single 2D images, we estimated 3D face reconstructions by fitting the morphable model. The reconstruction time measured on an Intel(R) Core (TM) Í5-8350U CPU @ 1.7 GHz was 58 minutes. We applied an approach proposed by Schönborn et al.23 to fit a 3DMM to a single 2D image. The fitting algorithm recovers a full posterior model of the face by simultaneously optimising facial shape and texture as well as illumination and camera parameters for a test face image. We used a spherical harmonics illumination model which can recover a broad range of natural illumination conditions in combination with a pinhole camera model. Illumination estimation is a critical step and optimised early and regularly in the sampling process. The fitting algorithm tries to reconstruct the 2D image, producing a rendering from the 3D model that matches the 2D image as closely as possible. The results for fitting a morphable model to a single 2D image are 3D face shape and texture reconstructions, as illustrated in the pipeline in Figure 2.
Experiments and results
Evaluating the face shape model
Before applying the 3DMM in our downstream reconstruction of a 3D face surface from a single 2D image, it was necessary to evaluate the quality of the built face model in terms of generalisation, specificity, and compactness. The details of the model evaluation metrics are discussed by Styner et al.33
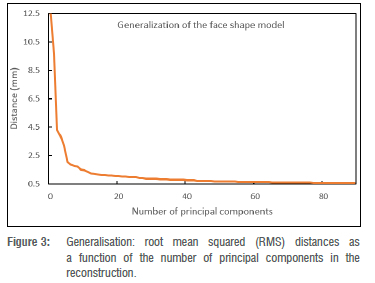
Shape model generalisation: This refers to the ability of the shape model to accurately represent an instance for which it was not trained. The leave-one-out approach33 was used to evaluate the generalisation ability of the face shape model. For each iteration, a shape model was constructed from a set of training face surfaces, leaving out one face shape instance. With all the training data in correspondence, the left-out face instance was projected into the shape model space to generate a face estimate. To evaluate the geometric accuracy of the estimated face, the distance between the face estimate and the original left-out face instance was calculated. The average vertex-to-vertex root mean squared (RMS) distance between the left-out face instance and the estimated face instance was computed. The procedure was repeated until all the face instances in the training set were used and each time the evaluation metric was calculated. The model generalisation ability results are presented in Figure 3, which demonstrates the generalisation error represented as RMS distance (y axis), plotted against shape principal components (x axis). After 5 principal components, the generalisation accuracy was close to 1.5 mm, and with 50 principal components, we reached an accuracy of approximately 0.5 mm.
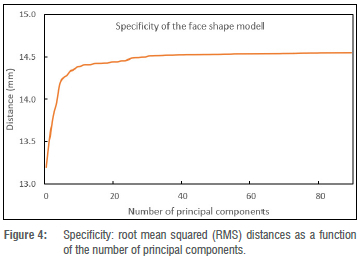
model specificity, a set of 90 shape instances was randomly generated from a distribution of the 3D morphable face model. The RMS distance between the randomly generated shape instances and the closest face surfaces in the training set was calculated as a specificity estimate. Lower RMS deviations are desirable because they indicate that the synthesised shape instances are close to the real shape instances in the training set. Figure 4 shows the specificity results. The results are in common ranges for specificity.15 Note that it is typical that the specificity decreases (distance increases) with greater model complexity (number of principal components).
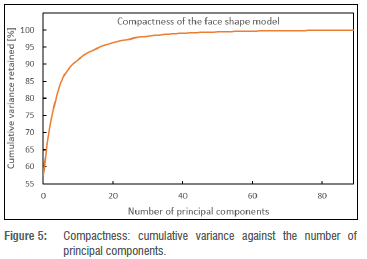
Shape model compactness: This indicates the percentage of variability accounted for by increasing numbers of principal components. Fewer principal components capture variability in shape information more efficiently. To validate the model compactness, the cumulative variance accounted for by the shape model was plotted as a function of the number of principal components of the shape model (illustrated in Figure 5). The line reflecting cumulative variance flattens as the number of shape principal components increases. Using only the first 20 shape principal components, the shape model accounts for more than 90% of shape variation in the training data set. This implies that the shape model is compact as it describes the training data set using a small number of principal components.
Evaluating 3D from 2D reconstruction results
To evaluate the geometric accuracy of face shape reconstructions from a single 2D image, the predicted 3D face surfaces were compared to the ground truth 3D face scans. We performed the reconstruction for each of the 98 2D images separately and build a separate 3DMM, removing that identity from the training data (leave-one-out cross-validation).
To measure the reconstruction error, we first rigidly aligned each predicted face mesh with the associated ground truth face shape. Following surface alignment, we computed the difference between the aligned face surfaces using the RMS distance metric.34 The RMS metric gives the surface-to-surface assessment value for each pair of surface comparisons. To visualise the reconstruction error distribution on the face surface, we additionally generated the surface colour maps from the comparisons of the predicted face surfaces and the associated ground truth face scans.
The overall average RMS error between the pairs of the predicted 3D face surfaces and the ground truth 3D face shapes is 2.62 mm with a deviation of 1.41 mm, with errors ranging from 1.00 mm to 6.75 mm. Furthermore, the visual shape comparisons between the predicted face surfaces and the corresponding ground truth face shapes are represented using colour gradients as illustrated in column (e) of Figure 6.
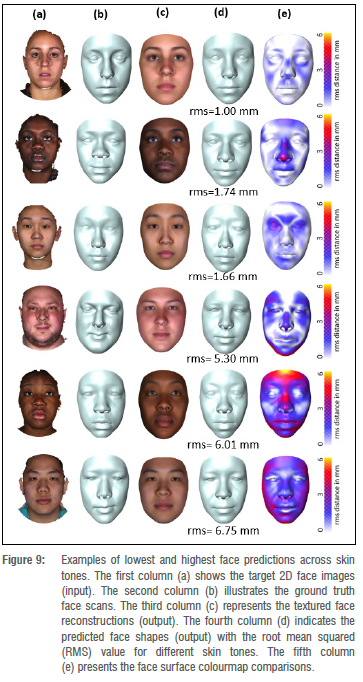
Figure 6 shows the identities with the best and worst RMS values for the predicted face shapes, and their corresponding ground truth face shapes, including the face surface colour map comparison. We observe that the largest reconstruction errors are found in regions of the face that are not relevant when screening for facial phenotypes in FAS. However, we find that the philtrum, which is one of the discriminators for FAS facial analysis, shows larger errors in faces considered to be outliers.
Face surface analysis across skin tones
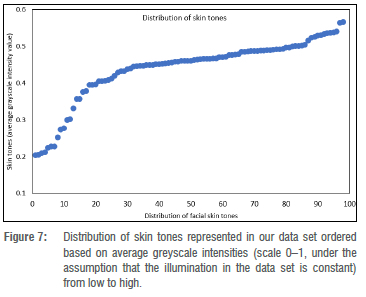
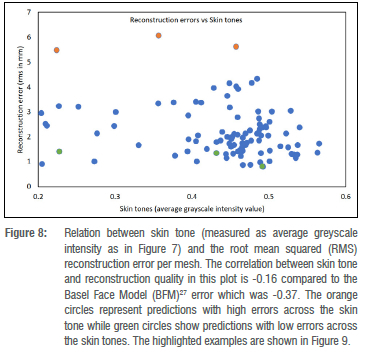
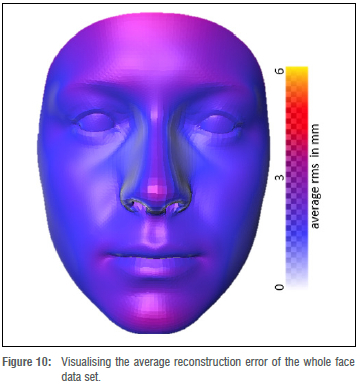
Foetal alcohol syndrome affects people of all ethnicities. Previous methods did not explore darker-skinned individuals well enough and structured light systems have acquisition issues when it comes to imaging darker skin tones.35 Furthermore, previous models have a strong bias towards lighter skin tones.36 We investigated what happens when darker and lighter skin tones are mixed. We can visualise the distribution of skin tones across our data set in Figure 7 and observe a heavy tail in the low intensity range. We also investigated the relationship between skin tone and the reconstruction error per mesh, and observe that we reach comparable reconstruction accuracy for the heavy tail of low intensity skin tones even though they are underrepresented in the training data (Figure 8). Figure 9 shows faces with lowest and highest reconstruction error results across skin tones. The poorest face reconstructions are also indicated in Figure 8 with orange circles, while the best face reconstructions are illustrated in the same figure with green circles. We find that regions of the face that are not related to the FAS facial phenotype are most affected. We also present the average reconstruction error over the whole data set in Figure 10.
3D surface distance measurements
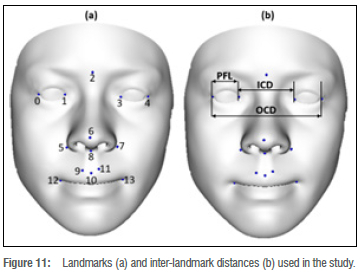
During FAS facial phenotype assessments, distances between facial features on the face surface can be measured using either a physical instrument or a computer-assisted tool. These surface measurements are used to confirm the diagnosis of facial syndrome. A study by Douglas et al.37 extracted facial features and performed measurements on the following face distances related to FAS: palpebral fissure length, inner canthal distance, outer canthal distance and interpupillary distance. However, these measurements were conducted on 2D stereo-photogrammetry images projected in 3D space. We extracted landmark points from our face reconstructions and can derive such measurements directly from our 3D reconstruction without manual interaction. We present reconstruction accuracy of those landmark points as well as distance reconstruction accuracy (measured in 3D) based on our 3D reconstructions compared to the ground truth 3D shapes.
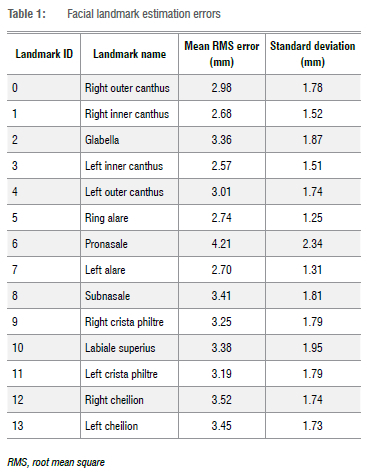
Landmark estimation. Landmarks are essential when taking measurements on a face surface. We identified and selected a subset of 14 landmark points which are related to FAS facial phenotype assessments. These landmarks are described in a study by Mutsvangwa and Douglas38. The results of the landmark estimation errors were computed and are illustrated in Table 1. The landmark error was computed by measuring position distance from reference surface to the reconstructed surface. As shown in Table 1,11 of 14 landmark errors were lower than 3.5 mm. The large standard deviations are mainly a result of the few outliers observed in Figure 8.
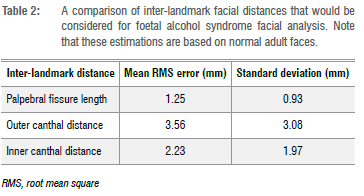
Facial feature distances. The distance measurements characteristic to FAS facial features include the palpebral fissure length, outer canthal distance, and inner canthal distance, as illustrated in Figure 11b. The landmarks required for the distance measurements are described in Table 1 and illustrated in Figure 11a. The corresponding distances on the reconstructed 3D face surface and the 3D ground truth face scan were compared and the difference calculated. Table 2 shows the results of average absolute distance errors and their standard deviations for the palpebral fissure length, outer canthal distance, and inner canthal distance facial feature distances.
Discussion
We constructed a 3D face model from 2D face scans and evaluated the accuracy of 3D face shape predictions from single images. The constructed morphable model of the face was evaluated for generalisation, specificity, and compactness parameters. The lowest generalisation error was 0.5 mm which suggests that the face shape model described the unseen face shapes well when given data outside the training set. The generalisation results of the face shape model compare well with other results found in the literature.15,16 The specificity results of the face shape model are in the range of 13.2 mm to 14.5 mm, which is in the common ranges for specificity.15 The compactness results of our face shape model indicate that more than 90% of the variability in the training set is retained with just 20 principal components and this compares well with other results in the literature. For example, Booth et al.16 report that the first 40 principal components retained more than 90% of variability in their training set. Overall, our morphable model construction and evaluation seem successful.
The numerical average reconstruction error between the reconstructed face shapes and the corresponding ground truth face shapes in our data set was 2.62 mm. These findings are comparable to other results in the literature. For example, Zollhofer et al.39 compared reconstructed face surfaces obtained from 3D face scans via a Kinect sensor to ground truth face scans, reporting an average deviation of about 2 mm. Additionally, Feng et al.40 reported a root mean square error of 2.83 mm from surface comparisons between the predicted 3D face meshes and the corresponding ground truth 3D face scans.
For FAS facial phenotype assessment, we are interested in specific regions of the face such as the eyes, the midface, the upper lip, and the philtrum. These regions provide cues to clinicians when examining the FAS facial phenotype. The whole face surface reconstruction was examined using the colourmap surface comparisons shown in Figures 6,9, and 10. From visual observation, the midface, the philtrum, and the regions around the eyes show lower levels of surface variation, as represented by the surface error colourmaps, whereas the upper lip areas show slight surface differences. The lower levels of surface variation around the eyes, the philtrum, and the midface could be attributed to the ease of identifying landmarks in those regions. From Figures 9 and 10, we observe that regions of the face which are not relevant to FAS facial phenotype analysis are most affected. A study by Hammond et al.41 suggested that visual inspections of the 3D surfaces using heat maps can delineate and discriminate facial features. In Figure 8, we find that the reconstruction quality in our data set is not affected by skin tones. On top of the heatmap representation, we also evaluated the landmarks and distances previously explored for the facial phenotype in FAS, as shown in Tables 1 and 2, respectively. We show accuracies in a minimum range of 2.57 mm for landmarking errors and 1.25 mm for distance errors. Similar results are reported in the literature. Regarding landmark localisation error, a study by Sukno et al.42 reported an average error per landmark of below 3.4 mm. For inter-landmark distances, Douglas et al.37 reported an average difference, between the manual and automated approaches, within 1 mm for palpebral fissure length, but with greater variations for outer canthal distance and inner canthal distance.
The highest face surface reconstruction errors belong to a relatively small set of 3D scans. Furthermore, the surface differences could imply that, during the model fitting phase of the reconstruction process, our statistical model did not completely capture all geometric cues in the 2D image of the face. We define a geometric cue as the information contained in a 2D image of the face, such as shading or contours.
Limitations and future research
Although we used a data set of scans of normal adult controls, with no known FAS indications, we assume that the framework is invariant to the data when built and applied to a population of interest. Ideally, training and test data sets would be collected from FAS and non-FAS control populations, with similar demographics. It is a challenge to access 3D data of individuals with FAS; however, the acquired face database (BU-3DFE) is very diverse. Future work could focus on reducing the reconstruction errors to acceptable clinical standards by collecting and analysing larger data sets, including more training data, especially from underrepresented populations. This would broaden the applicability of the morphable models of the face. To improve on the surface reconstruction performance, future developments could consider using multi-view 2D images of the face to provide more geometric cues during the model fitting of the face.
Conclusion
In this study, we aimed to evaluate whether an inverse graphics-based 3D from 2D reconstruction algorithm is suitable for acquiring 3D face data for FAS facial shape analysis. The reconstruction task was accomplished by fitting a 3DMM to a 2D image to recover a 3D face representation. Additionally, 3DMMs were built from a collection of 3D face scans with shape and texture information. We provided an evaluation performance of face reconstruction for future applications to FAS diagnosis. The resulting accuracies are promising for these future applications, even across different ethnicities.
Acknowledgements
This research was supported by the African Biomedical Engineering Mobility (ABEM) project, under the Intra-Africa Academic Mobility Scheme of the European Commission's Education, Audio Visual and Culture Executive Agency, and by the South African Research Chairs Initiative of the Department of Science and Innovation and the National Research Foundation of South Africa (grant no. 98788). B.E. is supported by a Postdoc Mobility Grant from the Swiss National Science Foundation (P400P2191110).
Competing interests
We have no competing interests to declare.
Authors' contributions
All authors participated in the project conceptualisation. FA. prepared the data, performed, and evaluated the experiments, and drafted the initial manuscript. B.E. performed and evaluated experiments and revised the manuscript. T.M. formulated the research goals and objectives, supervised the project, and provided financial support. All authors contributed towards the writing of the final manuscript.
References
1. Streissguth AR Bookstein FL, Barr HM, Sampson PD, O'Malley K, Young JK. Risk factors for adverse life outcomes in fetal alcohol syndrome and fetal alcohol effects. J Dev Behav Pediatr. 2004;25(4):228-238. https://doi.org/10.1097/00004703-200408000-00002 [ Links ]
2. Astley SJ, Clarren SK. Afetai alcohol syndrome screening tool. Alcohol Clin Exp Res. 1995;19(6):1565-1571. https://doi.Org/10.1111/j.1530-0277.1995.tb01025.x [ Links ]
3. Hoyme HE, May PA, Kalberg WO, Kodituwakku R Gossage JR Trujillo PM; et al. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: Clarification of the 1996 Institute of Medicine criteria. Pediatrics. 2005;115(1 ):39-47. https://doi.Org/10.1542/peds.2004-0259 [ Links ]
4. Clarren SK, Smith DW. The fetal alcohol syndrome. N Engl J Med. 1978;298(19):1063-1067. https://doi.Org/10.1056/NEJM197805112981906 [ Links ]
5. Astley SJ, Clarren SK. A case definition and photographic screening tool for the facial phenotype of fetal alcohol syndrome. J Pediatr. 1996;129(1):33-41. https://doi.Org/10.1016/S0022-3476(96)70187-7 [ Links ]
6. Astley SJ, Clarren SK. Diagnosing the full spectrum of fetal alcohol-exposed individuals: Introducing the 4-digit diagnostic code. Alcohol Alcohol. 2000;35(4):400-410. https://doi.Org/10.1093/alcalc/35.4.400 [ Links ]
7. Moore ES, Ward RE, Jamison PL, Morris CA, Bader PI, Hall BD. The subtle facial signs of prenatal exposure to alcohol: An anthropometric approach. J Pediatr. 2001 ;139(2):215-219. https://doi.org/10.1067/mpd.2001.115313 [ Links ]
8. May PA, De Vries MM, Marais A-S, Kalberg WO, Adnams CM, Hasken JM, et al. The continuum of fetal alcohol spectrum disorders in four rural communities in South Africa: Prevalence and characteristics. Drug Alcohol Depend. 2016;159:207-218. https://doi.Org/10.1016/j.drugalcdep.2015.12.023 [ Links ]
9. Honrado CR Larrabee WF Jr. Update in three-dimensional imaging in facial plastic surgery. Curr Opin Otolaryngol Head Neck Surg. 2004;12(4):327-331. https://doi.org/10.1097/01.moo.0000130578.12441.99 [ Links ]
10. Lekakis G, Claes R Hamilton GS, Hellings PW. Three-dimensional surface imaging and the continuous evolution of preoperative and postoperative assessment in rhinoplasty. Facial Plast Surg. 2016;32:88-94. https://doi.org/10.1055/S-0035-1570122 [ Links ]
11. Blanz V Vetter T. Face recognition based on fitting a 3D morphable model. IEEE Trans Pattern Anal Mach Intell. 2003;25(9):1063-1074. https://doi.org/10.1109/TPAMI.2003.1227983 [ Links ]
12. Egger B, Smith WA, Tewari A, Wuhrer S, Zollhoefer M, Beeler T, et al. 3D morphable face models - past, present, and future. ACM Trans Graph. 2020;39(5):1-38. https://doi.org/10.1145/3395208 [ Links ]
13. Blanz V Vetter T. Morphable model for the synthesis of 3D faces. In: Proceedings of the ACM SIGGRAPH Conference on Computer Graphics. New York: ACM Press/Addison-Wesley Publishing Co.; 1999. p. 187-194. https://doi.org/10.1145/311535.311556 [ Links ]
14. Booth J, Antonakos E, Ploumpis S, Trigeorgis G, Panagakis Y, Zafeiriou S, editors. 3D face morphable models "in-the-wild". In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2017. p. 48-57. https://doi.org/10.1109/CVPR.2017.580 [ Links ]
15. Dai H, Pears N, Smith WA, Duncan C, editors. A 3D morphable model of craniofacial shape and texture variation. In: Proceedings of the IEEE International Conference on Computer Vision. IEEE; 2017. p. 3085-3093. https://doi.org/10.1109/ICCV.2017.335 [ Links ]
16. Booth J, Roussos A, Ponniah A, Dunaway D, Zafeiriou S. Large scale 3D morphable models. Int J Comput Vision. 2018;126(2^1):233-254. https://doi.org/10.1007/s11263-017-1009-7 [ Links ]
17. Weiwei Y, editor. Fast and robust image registration for 3D face modeling. In: 2009 Asia-Pacific Conference on Information Processing; 2009 July 18-19; Shenzhen, China. IEEE; 2009. p. 23-26. https://doi.org/10.1109/APCIP.2009.142 [ Links ]
18. Amberg B, Romdhani S, Vetter T, editors. Optimal step nonrigid ICP algorithms for surface registration. In: 2007 IEEE Conference on Computer Vision and Pattern Recognition; 2007 June 17-22; Minneapolis, MN, USA. IEEE; 2007. p. 1-8. https://doi.org/10.1109/CVPR.2007.383165 [ Links ]
19. Jolliffe IT, Cadima J. Principal component analysis: A review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374(2065). Art. #20150202. https://doi.org/10.1098/rsta.2015.0202 [ Links ]
20. Romdhani S, Vetter T. Estimating 3D shape and texture using pixel intensity edges, specular highlights, texture constraints and a prior. In: 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05); 2005 June 20-25; San Diego, CA, USA. IEEE; 2005. p. 986-993. [ Links ]
21. Paysan P, Knothe R, Amberg B, Romdhani S, Vetter T. A 3D face model for pose and illumination invariant face recognition. In: 2009 Sixth IEEE International Conference on Advanced Video and Signal Based Surveillance. 2009;70(80):296-301. https://doi.org/10.1109/AVSS.2009.58 [ Links ]
22. Aldrian O, Smith WA. Inverse rendering of faces with a 3D morphable model. IEEE Trans Pattern Anal Mach Intell. 2013;35(5):1080-1093. https://doi.org/10.1109/TPAMI.2012.206 [ Links ]
23. Schônborn S, Egger B, Morel-Forster A, Vetter T. Markov chain Monte Carlo for automated face image analysis. Int J Comput Vision. 2016;123(2):160183. https://doi.Org/10.1007/S11263-016-0967-5 [ Links ]
24. Sanyal S, Bolkart T, Feng H, Black MJ, editors. Learning to regress 3D face shape and expression from an image without 3D supervision. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition: 2019 June 15-20; Long Beach, CA, USA. IEEE; 2019. p. 7763-7772. https://doi.org/10.1109/CVPR.2019.00795 [ Links ]
25. Feng Y, Feng H, Black MJ, Bolkart T. Learning an animatable detailed 3D face model from in-the-wild images. ACM Trans Graph. 2021 ;40(4), Art. #88. https://doi.org/10.1145/3450626.3459936 [ Links ]
26. Yin L, Wei X, Sun Y, Wang J, Rosato MJ, editors. A 3D facial expression database for facial behavior research. In: 7th International Conference on Automatic Face and Gesture Recognition (FGR06); 2006 April 10-12: Southampton, UK. IEEE; 2006. p. 211-216. [ Links ]
27. Gerig T, Morel-Forster A, Blumer C, Egger B, Luthi M, Schônborn S, et al., editors. Morphable face models - an open framework. In: 13th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2018); 2018 May 15-19; Xi'an, China. IEEE; 2018. p. 75-82. https://doi.org/10.1109/FG.2018.00021 [ Links ]
28. Lüthi M, Forster A, Gerig T, Vetter T. Shape modeling using gaussian process morphable models. In: Zheng G, Li S, Székely G, editors. Statistical shape and deformation analysis: London: Academic Press; 2017. p. 165-191. https://doi.org/10.1016/B978-0-12-810493-4.00008-0 [ Links ]
29. Lüthi M, Gerig T, Jud C, Vetter T. Gaussian process morphable models. IEEE Trans Pattern Anal Mach Intell. 2017;40(8):1860-1873. https://doi.org/10.1109/TPAMI.2017.2739743 [ Links ]
30. Gower JC. Generalized Procrustes analysis. Psychometrika. 1975;40(1):33-51. https://doi.org/10.1007/BF02291478 [ Links ]
31. Goodall C. Procrustes methods in the statistical-analysis of shape. J Roy Stat Soc B Met. 1991 ;53 (2) :285-339. https://doi.org/10.1111/J.2517-6161.1991.tb01825.x [ Links ]
32. Komalasari D, Widyanto MR, Basaruddin T, Liliana DY, editors. Shape analysis using generalized Procrustes analysis on active appearance model for facial expression recognition. In: 2017 International Conference on Electrical Engineering and Computer Science (ICECOS); 2017 August 22-23; Palembang, Indonesia. IEEE; 2017. p. 154-159. https://doi.org/10.1109/ICECOS.2017.8167123 [ Links ]
33. Styner MA, Rajamani KT, Nolte L-P, Zsemlye G, Székely G, Taylor CJ, et al. Evaluation of 3D correspondence methods for model building. In: Taylor C, Noble JA, editors. Information Processing in Medical Imaging (IPMI 2003). Lecture Notes in Computer Science vol 2732. Berlin: Springer; 2003. p. 63-75. https://doi.org/10.1007/978-3-540-45087-0_6 [ Links ]
34. Chai T, Draxler RR. Root mean square error (RMSE) or mean absolute error (MAE)? - arguments against avoiding RMSE in the literature. Geosci Model Dev. 2014;7(3):1247-1250. https://doi.org/10.5194/gmd-7-1247-2014 [ Links ]
35. Voisin S, Page DL, Foufou S, Truchetet F, Abidi MA. Color influence on accuracy of 3D scanners based unstructured light. In: Meriaudeau F, Niel KS; editors. Proceedings volume 6070 Machine Vision Applications in Industrial Inspection XIV. SPIE; 2006, #607009. https://doi.Org/10.1117/12.643448 [ Links ]
36. Grother P, Ngan M, Hanaoka K. Face recognition vendor test (FRVT). Part 2: Identification. Gaithersburg, MD: US Department of Commerce, National Institute of Standards and Technology; 2019. https://doi.org/10.6028/NIST.IR.8271 [ Links ]
37. Douglas TS, Martinez F, Meintjes EM, Vaughan CL, Viljoen DL. Eye feature extraction for diagnosing the facial phenotype associated with fetal alcohol syndrome. Med Biol Eng Comput. 2003;41 (1):101-106. https://doi.org/10.1007/BF02343545 [ Links ]
38. Mutsvangwa T, Douglas TS. Morphometric analysis of facial landmark data to characterize the facial phenotype associated with fetal alcohol syndrome. J Anat. 2007;210(2):209-220. https://doi.Org/10.1111/J.1469-7580.2006.00683.x [ Links ]
39. Zollhofer M, Martinek M, Greiner G, Stamminger M, Sussmuth J. Automatic reconstruction of personalized avatars from 3D face scans. Comput Animation Virtual Worlds. 2011 ;22(2-3):195-202. https://doi.org/10.1002/cav.405 [ Links ]
40. Feng Z-H, Huber P, Kittler J, Hancock P, Wu X-J, Zhao Q, et al., editors. Evaluation of dense 3D reconstruction from 2D face images in the wild. In: 13th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2018); 2018 May 15-19; Xi'an, China. IEEE; 2018. p. 780-786. https://doi.org/10.1109/FG.2018.00123 [ Links ]
41. Hammond P, Hutton TJ, Allanson JE, Buxton B, Campbell LE, Clayton-Smith J, et al. Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet. 2005;77(6):999-1010. https://doi.org/10.1086/498396 [ Links ]
42. Sukno FM, Waddington JL, Whelan PF. 3-D facial landmark localization with asymmetry patterns and shape regression from incomplete local features. IEEE Trans Cybern. 2014;45(9):1717-1730. https://doi.org/10.1109/TCYB.2014.2359056 [ Links ]
 Correspondence:
Correspondence:
Tinashe Mutsvangwa
Email: tinashe.mutsvangwa@uct.ac.za
Received: 24 Aug. 2021
Revised: 22 Feb. 2022
Accepted: 23 Nov. 2022
Published: 29 Mar. 2023
Editor: Michael Inggs
Funding: European Commission; South African Department of Science anc Innovation; South African National Research Foundation (grant no. 98788); Swiss National Science Foundation (P400P2J91110)














