Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.118 no.9-10 Pretoria sep./oct. 2022
http://dx.doi.org/10.17159/sajs.2022/12590
COMMENTARY
Cautioning the move from morphology to molecules in the taxonomy of Metazoa: Comments on Lawley et al. (PeerJ 2021;9, e11954) and a plea for considered integration
Michael K. Brown; Mark J. Gibbons
Department of Biodiversity and Conservation Biology, University of the Western Cape, Cape Town, South Africa
ABSTRACT
SIGNIFICANCE: This paper serves as a commentary on a recently published paper by Lawley et al. (PeerJ 2021;9, e11954). We caution the adoption of practices in the taxonomy of Scyphozoa by Lawley et al. on the basis that they may lead to taxonomic splits and parallel taxonomies in the face of a concerted push towards integrative taxonomy
Keywords: integrative taxonomy, molecular species description, morphological species description, crypsis
Species are such fundamental and important units that they should not be introduced carelessly. Species description and splitting based on superficial data like simple morphometric differences (including those that are statistically significant), arbitrary values of genetic distance or phylogenetic relationships derived from limited molecular datasets (single-locus analyses, particularly mtDNA) is strongly discouraged. All of these may serve to support conclusions derived from more appropriate datasets, but are not sufficient on their own.1
Lawley et al.2 recently erected/resurrected a number of new species of Scyphozoa in the genus Aurelia, on the basis of molecular markers alone. They took this approach because, while morphological data were effectively absent for some, the genus is characterised by morphological crypsis. Although the arguments advanced by Lawley et al.2 provide us with an opportunity to discuss alternative methods of taxonomy in Scyphozoa, we caution against their immediate adoption by the wider community, as they potentially serve to create chaos and instability.
In their Introduction, Lawley et al.2 comprehensively articulate the well-understood problems of crypsis in the genus Aurelia, and their results highlight the issue of ecologically driven morphoplasticity, which serves to further complicate the taxonomic task at hand. Although Lawley et al.2 did not dismiss an integrative approach to taxonomy, which relies on congruence in multiple lines of investigation before species descriptions are drafted, they chose rather to rely on a single method - the use of an often-incomplete suite of molecular markers (16S, COI, ITS1, 28S; see Table 1).
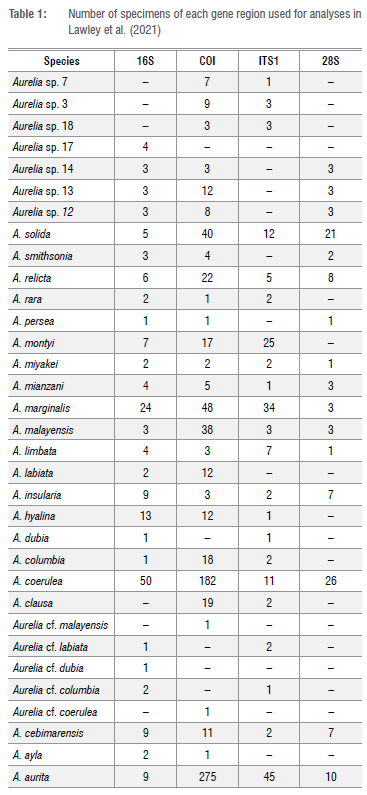
Lawley et al.'s2 decisions about species identity were based in part on species distributions and in part on the topology, synapomorphies and support of concatenated phylogenetic trees. While the trees for individual genetic markers were included in supplementary material, their results were not discussed in the paper. They ignored levels of genetic divergence, arguing that while such may provide 'a useful tool for first assessments and the discovery of potentially cryptic species, …. it might not be reliable for species identification…[and should not be used]… for species delimitation.' We accept this logic, given that '….evolutionary rates may vary across congeners…[and that]…similarity does not necessarily reflect kinship….'. However, we are worried that some may see the 2% divergence between sister taxa as a flag for the erection of species in instances where fuller data are missing. Such could result in a flurry of spurious (potentially artefactual) new species descriptions based on single specimens. While it may have been acceptable in the 19th century to describe a species based on a single specimen (e.g. Cyanea annasethe3), that approach can no longer be appropriate unless the specimen being described appears so obviously different from known diversity (e.g. Chirodectes maculatus4), given what we now know about genetic variability within a single species from even one location. Many recent taxonomists have spent a great deal of time and effort describing and validating just one new species in a genus from multiple specimens in order to incorporate variability (e.g. Scorrano et al5; Ras et al.6). So, it is with some concern that we note Lawley et al.2 have increased the number of species in the genus Aurelia from 19 to 297 on such a scant basis in some instances (Table 1).
It should be a given that new molecular species descriptions will need to be rigorously executed and more rigorously reviewed, but we need to recognise that even the most thorough of descriptions may in fact be guilty of renaming an already valid species, originally defined by morphological taxonomy, but for which corresponding molecular material is missing. While renaming an already valid species is not new in taxonomy, it is a practice that needs to be avoided as it sows confusion. Aurelia solida is a case in point. The species was first described by Browne from a single specimen collected around the Maldives, and although it has never been recovered from the type locality since, it has been recorded from the Mediterranean and Red Seas subsequently.5 Designation of the species in the latter instances was based primarily on morphological grounds, with molecular data serving to confirm its unique identity.5 Although it has been hypothesised that the species was moved from the Indian Ocean into the Mediterranean Sea via ships8, it is equally possible that the Type specimen and the material collected from the Mediterranean are in fact different. Given this, and given that multiple species of Aurelia may occur in sympatry (Figure 10 in Lawley et al.2), it becomes very clear that robust conclusions about identity are hard to reach. An observation that in this instance is made more pertinent by the fact that another 'never-again' caught species was first collected from the Type locality: Aurelia maldivensis. Which naturally leads us to question Lawley et al.'s2 use of sample locality as a proxy for species grouping.
Despite concluding that there is a need for us to understand the processes that lead to morphological variation in Aurelia, Lawley et al.2 effectively question its value in taxonomy. How then do we address species descriptions within the genus that were published before the advent of molecular analysis? If the argument is that Aurelia exhibit far too much morphoplasticity in response to ecological processes, the implication is that we will never be able to resurrect, or confirm, species descriptions based on morphological descriptions in published works. This means that we could end up with two parallel, non-overlapping taxonomies, which will confuse rather than clarify - unless we simply declare many of the old names species inquirendae and consign them to the dustbin of history.
If the approach proposed here is widely adopted, it is not impossible to imagine fleets of autonomous underwater vehicles (AUVs) moving around the world's oceans throwing out new species descriptions on a regular basis using on-board molecular technologies that link to land-based supercomputers by satellite feeds. Such would indeed help us to understand molecular diversity at the global level, and would help us in real-time monitoring using e-DNA, but we wonder whether it helps us to understand evolution. We believe that the solution to this is clear. Integrative taxonomy. Reserving full description until fuller data across all aspects of morphology, cnidome, ecology and DNA (especially population genetics, which must be the underlying basis for all objective species limitations), etc. are obtained (as Dawson9). While there is a need for us to understand and manage global biodiversity before it is lost, we do not need to conflate that task with taxonomy. We therefore reiterate our call for caution in the adoption of Lawley et al.'s2 approach.
Acknowledgements
We thank Michael Dawson, Nando Boero and Stefano Piraino for their thoughtful comments on the manuscript, which have given us confidence in our arguments. We also extend our gratitude to the three anonymous readers, whose helpful and positive feedback has been incorporated into a revised text. The editorial team at the South African Journal of Science is thanked for giving us space to publish our response to a paper published in PeerJ. We thank the National Research Foundation and the University of the Western Cape for ongoing financial and logistical support.
References
1.Zachos FE. Tree thinking and species delimitation: Guidelines for taxonomy and phylogenetic terminology. Mamm Biol. 2016;81:185-188. https://doi.org/10.1016/j.mambio.2015.10.002 [ Links ]
2.Lawley JW, Gamero-Mora E, Maronna MM, Chiaverano LM, Stampar SN, Hopcroft RR, et al. The importance of molecular characters when morphological variability hinders diagnosability: Systematics of the moon jellyfish genus Aurelia (Cnidaria: Scyphozoa). PeerJ. 2021;9, e11954. https://doi.org/10.7717/peerj.11954 [ Links ]
3.Haeckel E. Das System der Medusen. pt. 2, System der Acraspeden [The system of the medusas. Part 2: System of acraspedes]. Monographie der Medusen 1880 1. G. Fischer, Jena. German. [ Links ]
4.Cornelius PFS, Fenner PJ, Hore R. Chiropsalmus maculatus sp. nov., a cubomedusa from the Great Barrier Reef. Mem Queensl Mus. 2005;51:399-405. [ Links ]
5.Scorrano S, Aglieri G, Boero F, Dawson MN, Piraino S. Unmasking Aurelia species in the Mediterranean Sea: An integrative morphometric and molecular approach. Zool J Linn Soc. 2016;180(2):243-267. https://doi.org/10.1111/zoj.12494 [ Links ]
6.Ras V, Neethling S, Engelbrecht A, Morandini AC, Bayha KM, Skrypzeck H, et al. There are three species of Chrysaora (Scyphozoa: Discomedusae) in the Benguela Upwelling Ecosystem, not two. Zootaxa. 2020;4778(3):401-438. https://doi.org/10.1111/zoj.12494 [ Links ]
7.World Register of Marine Species (WoRMS) [homepage on the Internet]. No date [cited 2021 Sep 14]. Available from: http://www.marinespecies.org [ Links ]
8.Gueroun SM, Molinero JC, Piraino S, Dali Yahia MN. Population dynamics and predatory impact of the alien jellyfish Aurelia solida (Cnidaria, Scyphozoa) in the Bizerte Lagoon (southwestern Mediterranean Sea). Mediterr Mar Sci. 2020;21(1):22-35. https://doi.org/10.12681/mms.17358 [ Links ]
9.Dawson MN. Renaissance taxonomy: Integrative evolutionary analyses in the classification of Scyphozoa. J Mar Biol Assoc UK. 2005;85(3):733-739. https://doi.org/10.1017/S0025315405011641 [ Links ]
 Correspondence:
Correspondence:
Michael Brown
Email: 3650408@myuwc.ac.za
Published: 29 September 2022














