Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.118 n.1-2 Pretoria Jan./Feb. 2022
http://dx.doi.org/10.17159/sajs.2022/10693
RESEARCH ARTICLE
Antiviral activity of chitosan nanoparticles for controlling plant-infecting viruses
Ahmed Y El GamalI; Mahmoud M. AtiaII; Tarek El SayedI; Mohamed I. Abou-ZaidII; Mohamed R. TohamyII
IVirus and Phytoplasma Research Department, Plant Pathology Research Institute, Agricultural Research Center, Giza, Egypt
IIPlant Pathology Department, Zagazig University, Zagazig, Egypt
ABSTRACT
Chitosan nanoparticles (ChiNPs) are a potentially effective means for controlling numerous plant diseases. This study firstly describes the antiviral capabilities of ChiNPs to control plant viral diseases compared to its bulk form. Bean yellow mosaic virus (BYMV) was used as a model plant virus affecting faba bean plants and many other legumes. The antiviral effectiveness of ChiNPs and chitosan were evaluated as a curative application method, using six dosage rates (50, 100, 200, 250, 300 and 400 mg/L). Results indicated that ChiNPs curatively applied 48 h post virus inoculation entirely inhibit the disease infectivity and viral accumulation content at 300 mg/L and 400 mg/L. The virus titre was greatly alleviated within the plant tissues by 7.71% up to100% depending on ChiNP dosage rates. However, chitosan used in its bulk-based material form revealed a relatively low to an intermediate reduction in virus infectivity by 6.67% up to 48.86%. Interestingly, ChiNPs affect the virus particle's integrity by producing defective and incomplete BYMV viral particles, defeating their replication and accumulation content within the plant tissues. Simultaneously, ChiNP applications were appreciably shown to promote the pathogenesis-related (PR-1) gene and other defence-related factors. The mRNA of the PR-1 gene was markedly accumulated in treated plants, reaching its maximum at 400 mg/L with 16.22-fold relative expression change over the untreated control. Further, the total phenol dynamic curve was remarkably promoted for 30 days in response to ChiNP application, as compared to the untreated control. Our results provide the first report that chitosan-based nanomaterials have a superior effect in controlling plant viruses as an antiviral curing agent, suggesting that they may feasibly be involved in viral disease management strategies under field conditions without serious health concerns and environmental costs.
SIGNIFICANCE:
• Our findings show that chitosan nanoparticles have a powerful curing antiviral activity against BYMV disease. These findings open the door for the use of eco-friendly nano-based tools in controlling numerous plant viruses. The use of eco-friendly nano-based materials could result in a successful integrative control strategy for plant viruses under field conditions, negating the need for the conventional measure used to control most of the insect-transmitted plant viruses, that is insecticide application against vector insects.
Keywords: chitosan nanoparticles, Bean yelbw mosaic virus (BYMV), antivirus, faba bean, PR-1 gene regulation
Introduction
Plant viruses are destructive diseases that cause serious concerns for the agroecosystem and global food security due to their ability to infect many plant species that are grown for both food and feed production.1 Faba bean (Vicia faba L.) is a legume crop cultivated mainly for its edible seeds. Due to their high protein content, faba beans are widely used as a cheap and high-quality protein source for poor consumers in many developing countries.2Bean yellow mosaic virus (BYMV) belongs to the Potyvirus genus (Family; Potyviridiae) and is a devastating viral disease for many crop legumes and ornamental plants throughout the world.3 BYMV disease poses a significant threat for faba bean production, with losses of 30-50%.4 However, under severe epidemic conditions, the disease can cause a total crop failure.5 The virus is naturally transmitted by over 20 species of aphids, resulting in a high rate of viral infections for faba bean and other host species.6 BYMV and most plant viruses are conventionally managed by pesticide applications against insect vectors.7 However, from a sustainability point of view, the extensive use of pesticides tremendously affects the environment and the whole ecosystem. According to the World Health Organization, thousands of agricultural workers in developing countries die each year from severe poisoning by pesticides.8 Therefore, new strategic technologies are urgently needed to avoid pesticide treadmills.
Nanotechnology-based tools have provided great hope for scientific revolutions in the near-future in many applications.9 The engineered nanoparticles have superior chemical and physical features compared to their bulk materials, allowing them to be used in many different sectors.10 Chitosan (poly (1,4)"2"amino"2"deoxy"p"D glucose) is a glucosamine polymer obtained by alkaline deacetylation of chitin extracted from the exoskeleton of crustaceans.11 From an ecological point of view, chitosan has received considerable interest due to its potential wide applications in plant protection and growth promotion because of its excellent biodegradability, non-toxicity and bioactivity.12'13 Moreover, chitosan has unique antimicrobial properties and it can additionally be used as an elicitor molecule against different plant pathogens and viral infections when used preventatively before inoculation.14-17 In limited previous studies, the partial antiviral potential of chitosan bulk material in controlling some plant viruses has been investigated for use of chitosan as a protective agent involved in the stimulation of plant defence systems against virus invasion.1819
In addition to the capability of the chitosan polymer for use preventatively as a plant defence enhancer against pathogen attacks, it has since been found that this capability is further enhanced by using it in the form of nanoparticles.20 This study was performed to evaluate the antiviral capability of the nanoparticle-based form of chitosan against plant virus infectivity in comparison with that of the chitosan bulk polymer. We also investigated their effects on some defence-related parameters involved in plant immunity mechanisms when curatively applied after a viral challenge in faba bean plants.
Materials and methods
Virus isolation and propagation
Faba bean plant samples with Bean yellow mosaic virus-like symptoms were collected from local faba bean fields in the Giza governorate, Egypt, during the 2017/2018 growing season. Leaf samples were initially tested using the double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) technique as described by Clark and Adams21 using specific polyclonal antibodies against BYMV (EPHYRA Bioscience Inc., Ontario, Canada). The reactions were assessed at 405 nm in a microplate reader (Bio-Tek, USA). Samples that tested positive against BYMV were used as sources of virus inoculum. The virus was isolated through single local lesion inoculations on Chenopodium amaranticolor indicator plants.22 Ten-day-old plants were mechanically inoculated with the BYMV crude sap using 0.01 M phosphate buffer pH 7.1, (1:10), 0.01% Na2SO3 and carborundum (600 mesh) and kept under insect-proof greenhouse conditions until symptoms developed. The isolated source was propagated on 10-day-old faba bean plants (cv. Giza 843) and was used as inoculum for further studies.
Molecular identification of the isolated virus
Total RNA was isolated from both healthy and symptomatic faba bean leaves using RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. Reverse transcription (RT)-PCR reaction was optimised using the One-Step RT-PCR system (Thermo Fisher, USA).
A specific primer pair, BYMV-CPU (5' GTCGATTTCAATCCGAACAAG 3') and BYMV-CPD (5' GGAGGTGAAACCTCACTAATAC 3'), was used to target the CP gene region to amplify a 907-bp fragment of the BYMV genome.23 The one-step RT-PCR reaction was performed by combining 25 ÄL One-Step PCR Master Mix, 1 ÄL of each primer pair (200 nM), 2.5 ÄL of RT-enzyme enhancer, 1 ÄL verso enzyme mix, and 3 ng of RNA template, and the mixture was made up to 50 ÄL using nuclease-free water. The amplification reaction was automated in a T-Gradient thermal cycler (Biometra, Germany) with an initial reverse transcription process at 50 °C for 15 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 2 min, with a final additional extension step for 10 min at 72 °C. The PCR products were electrophoresed on 1% agarose gel in 0.5 x TAE buffer, then visualised with a Gel Doc system (2000, Bio-Rad, USA).
Preparation of chitosan nanoparticles
A chitosan nanoparticle solution was chemically prepared by reduction of low molecular weight chitosan (Sigma, Egypt) based on the ion-gelation method using sodium tripolyphosphate as a reducing agent.24 Chitosan powder (0.5 mg/mL) was dissolved in 1% acetic acid in deionised distilled water and left under vigorous stirring for 30 min. Sodium tripolyphosphate was dissolved separately in deionised distilled water (0.7 mg/mL). Chitosan nanoparticles were formed by mixing 500 mL of chitosan with increasing amounts of sodium tripolyphosphate solution (160 mL) under continuous stirring for 1 h. The synthesised chitosan nanoparticles (ChiNPs) were subjected to further physicochemical characterisation.
Physicochemical characterisation of ChiNPs
Dynamic light scattering analysis
The particle size distribution and zeta potential of constructed ChiNPs were assessed using a Zetasizer (Malvern, ZS Nano, UK). The colloidal chitosan nanoparticle solution was diluted with distilled water and put into a disposable cuvette for analysis.
X-ray diffraction
The physicochemical crystalline nature of ChiNPs was confirmed using an X-ray diffractometer (Xpert PRO, PAN analytical, Netherlands) operated with a CuK radiation tube (= 1.54 A°) at 40 kV. The ChiNP solution was centrifuged at 20 000 x g for 30 min at 4 °C for powder phase yield, the precipitated ChiNPs were dried, then bombarded by X-ray for phase analysis.25
Surface morphology
Particle size and the actual shape of ChiNPs were determined by high-resolution transmission electron microscope (Tecnai G2, FEI, Netherlands) under an accelerating voltage of 200 kV.
Effect of ChiNPs on virus infectivity
Treatments
Foliar applications were carried out under an insect-proof greenhouse using six concentrations of chitosan and ChiNPs (50, 100, 200, 250, 300 and 400 mg/L). Ten-day-old faba bean plants (five plants/pot) were mechanically inoculated with BYMV- infected plant sap. The inoculated plants were uniformly sprayed until runoff with all tested dosage rates (48 h post-viral challenge). Water-treated plants served as a comparable control. Each experimental group had four replicates.
Disease assessment
Virus infectivity was determined 3 weeks post-inoculation on all treated and untreated faba bean plants using the assessment of disease incidence and severity response. Disease severity and symptom response were also assessed using a numerical scale of grades 0-4, where 0=no visible symptom apparent; 1=mild chlorotic patterns; 2=mosaic patterns and dark green vein banding; 3=mosaic patterns, leaf distortion, and reduction in leaf size; 4=severe mosaic and stunting of the whole plant. Values of disease severity were estimated by the following formula26:
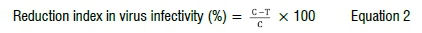
The inhibition index in virus infectivity was calculated using the following formula:

where C is the mean average of virus incidence values in untreated control plants; and T is the average value in each treatment.
Transmission electron microscope
For electron microscopic examination of virus particles, the leaf-dip preparation method was performed on the faba bean plants treated with 400 mg/L of ChiNPs and untreated controls 5 days post-treatment. Briefly, the leaf samples of both treated and untreated plants were washed, a small disc was prepared (1.5 cm in diameter) and resuspended in deionised water to remove any surface-ChiNPs attached to the leaf samples. The samples were ground in a drop of phosphate buffer pH 7.5 and 0.01% Na2SO3. Leaf extracts (10 ÄL) were individually loaded on carbon-coated grids for 5 min, washed with distilled water and negatively stained with 2% uranyl acetate.27 The grids were examined using a JEOL JEM1400 transmission electron microscope (JEOL Co., Tokyo, Japan).
Determination of virus accumulation content
Newly emerged small leaves were collected 30 days post-inoculation to detect the BYMV accumulation and to further confirm the virus replication inhibition rate using the DAS-ELISA method.21 The virus titre reactions were quantified at 405 nm in a microplate reader (BioTek, USA). Samples were considered positive when optical density (0D)-405 values were two times higher than the mean of the healthy control. The reduction percentage in virus accumulation was measured as follows:
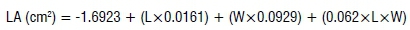
where Vt.A is the mean average of virus titre OD values in positive untreated control plants and Vt.B is the average virus titre OD value in each ChiNP treatment.
Enzyme activity assays
Fresh leaves (0.2 g) from all treated and untreated plants were collected at 0, 24, 48, 72, 96, 120, and 144 h post-ChiNP spraying. The polyphenol oxidase antioxidant enzyme activity was determined using the methods described by Kar and Mishra.28 Change in activity was expressed as nmol of guaiacol per mg protein per min. Phenylalanine ammonia lyase activity was measured according to Lisker et al.29 Values are expressed as nmol of cinnamic acid/gfw/s. All experiments were performed in triplicate.
Pathogenesis-related (PR-1) gene expression analysis
Total RNA isolation
Leaf samples from ChiNP-treated and untreated control plants were collected 48 h post-treatment for total RNA isolation. The total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) protocol.
cDNA synthesis
The harvested RNA (2 Äg) was primed with Oligo (dT) and converted into the first-strand cDNA using COSMO cDNA synthesis kit (Willowfort, UK) according to the manufacturer's protocol. The cDNA synthesis reaction was performed in a T-Gradient thermal cycler with initial annealing at 25 °C for 10 min, followed by an extension phase at 45 °C for 15 min.
Primers used
Gene-specific primers encoding pathogenesis-related protein 1 (PR-1 forward 5'-GGGCAGTGGTGACATAACAGGAA-3') upstream and (PR-1 reverse 5'-CATCCAACCCGAACCGAAT-3') downstream were designed.30 A specific primer pair of the elongation factor 1-alpha gene ELF1A/ forward (5'-GTGAAGCCCGGTATGCTTGT-3') and ELF1 A/reverse (5'-CTTGAGATCCTTGACTGCAA CATT-3') was used as an endogenous reference gene.
qRT-PCR analysis
The HERA SYBR Green qPCR kit (Willowfort, UK) was used for qRTPCR analysis with a 20-μL reaction mixture consisting of 10 μL SYBR green master mix, nuclease-free water (7.2 μL), diluted cDNA template (2 μL) and 0.5 μL of each primer pair. The qRT-PCR programme was as follows: 95 °C for 2 min, followed by 40 cycles at 95 °C for 1 min and 60 °C for 30 s. The reaction was normalised with melting curve analysis at 65 °C for 10 s for 61 cycles. The changes in gene expression were calculated based on the internal reference gene using the 2-ΔΔCt method.31 Each experiment was conducted in triplicate.
Determination of total phenolic content
The total polyphenol dynamic curve was determined following the Folin-Ciocalteu's reagent according to the methods described by Lin and Tang32 with slight modifications. Briefly, the collected leaf samples (0.5 g) were vigorously homogenised in 10 mL absolute ethanol. The leaf extract solution (100 µL) was transferred in a test tube containing 4 mL distilled water and 1 mL of Folin-Ciocalteu's reagent. After 5 min, 1 mL of sodium bicarbonate (10%) and 1 mL of ethanolamine were added to the mixture. The tubes were shaken and left for 1 h in the dark at room temperature. The samples were electro-optically measured at 740 nm. A standard calibration curve for total phenol was established using gallic acid (0- 200 ppm). The results were expressed as mg of gallic acid equivalents/gram fresh weight (gfw).
Effects on plant growth and vegetation parameters
Vegetation and growth parameters (i.e. leaf area, shoot length and chlorophyll content) of treated and untreated faba bean plants were determined at the end of the experiment. Leaf area was estimated following the estimation model proposed by Chahit33. For each tested concentration, 20 leaflets of all treated plants were measured. The maximum length of the leaflet was excised from petiole to the tip along the mid-vein, while the width was obtained by measuring the area between two leaflet margins perpendicular to the mid-vein. The leaf area was estimated by:

where LA is leaf area in cm2, L is leaflet length (cm), and W is the leaflet width (cm).
Changes in chlorophyll content were automatically assessed using a portable chlorophyll meter (SPAD-502 Plus, Minolta UK) and the obtained results were expressed as a chlorophyll content index. Leaflets from all treated and untreated plants were assessed and chlorophyll index values were obtained for all tested concentrations. Finally, shoot length (in cm) was also measured for all treated and untreated plants from the soil line to the top of the plant using a one-metre tape measure.
Statistical analysis
Data were statistically analysed using a completely randomised design for analysis of variance (ANOVA). Statistical analysis of the data was performed with the Assistat-Statistics Software (version 7.7 beta).34 The significant differences in each treatment group were determined at p = 0.05. Each experiment was performed in triplicate.
Results
Virus isolation and characterisation
The biologically isolated source produced mosaic symptoms typical of BYMV disease and similar to those observed in the field (Figure 1a). BYMV isolate was confirmed by DAS-ELISA in mechanically inoculated plants and all tested samples were positive for BYMV. The virus titre was 1.113 OD in faba bean, compared with 0.230 OD for negative control plants. All tested samples failed to react with antiserum specific for other viruses affecting faba bean plants. The RT-PCR products for the CP gene of BYMV were confirmed with 1% agarose gel electrophoresis. Two clear bands of 907 bp, the expected amplicon size, were produced from BYMV-infected samples. However, no pCr products were achieved from healthy faba bean plants used as a negative control (Figure 1b).
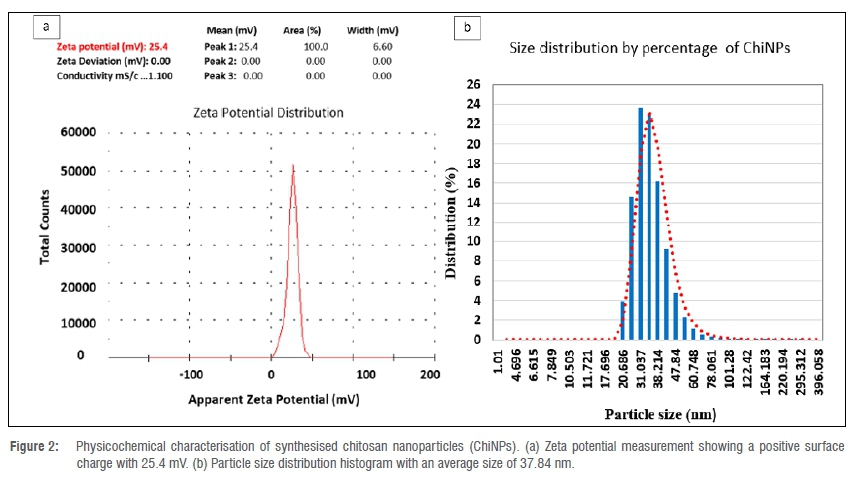
Characterisation of ChiNPs
The synthesised ChiNPs showed an average hydrodynamic size distribution of 37.84 nm (ranging between 20.68 nm and 60.74 nm) with a polydispersity index of 0.423 (Figure 2a). The zeta potential of ChiNPs indicates a positive surface area charge of 25.4 mV (Figure 2b). The amorphism fingerprint nature of ChiNPs is represented on the X-ray diffraction system by the formation of a hump shape from 10 to 30 2Θ angle (Figure 3a). The transmission electron micrograph illustrates a pseudospherical shape for the synthesised ChiNPs and the average particle size was estimated to be about 36.52 nm (Figure 3b).
Virucidal activity of ChiNPs on BYMV infectivity
ChiNPs suppressed BYMV replication on faba bean plants when compared to the bulk chitosan polymer form (Figure 4a,c). Untreated control plants infected with BYMV showed severe mosaic and stunting symptoms. Interestingly, symptoms were not observed with plants treated with 300 mg/L and 400 mg/L ChiNPs, with a complete reduction of the disease response 48 h post virus inoculation. Faba bean plants treated with 100, 200 and 250 mg/L ChiNPs revealed different levels of protection against BYMV occurrence of 47.67%, 43.34% and 75.40%, respectively. However, a low to moderate reduction in virus infectivity was obtained in bulk chitosan-treated plants - 21.11% and 48.86%, respectively - compared with the untreated control plants (Figure 4b,d).
In this respect, transmission electron micrographs illustrate that ChiNP applications resulted in defective and incomplete particles compared to the untreated control (Figure 5). The viral particles obtained from the ChiNP-treated plants were clearly found to be lower than 300 nm in length (Figure 5b,c,d). Contrastingly, the normal well-devolved virus particles in untreated control plants were estimated to be 715-730 nm in length (Figure 5a).
Effect of ChiNPs on virus accumulation content
ChiNPs appreciably decreased virus accumulation in treated faba bean plants to varying levels depending on the ChiNP dosage rate. All tested concentrations of ChiNPs, except for 50 mg/L and 100 mg/L, significantly reduced virus accumulation in the treated plants when applied 48 h postinoculation (Table 1). The highest activity was obtained when the plants were sprayed with 300 mg/L and 400 mg/L in which a negative ELISA reaction was observed with no significant difference compared to the negative control. A significant difference was recorded after treatment with 200 mg/L and 250 mg/L ChiNPs, with the virus content significantly decreased by 44.95% (0.915 OD) and 60.18% (0.649 OD), respectively, compared to the untreated control. However, bulk chitosan application showed that the BYMV accumulation content did not change significantly from all tested concentrations, only from 300 mg/L and 400 mg/L and the virus titre was significantly reduced by 36.12% (0.945 OD) and 42.66% (0.880 OD), respectively, as compared with the untreated control (1.244 OD).
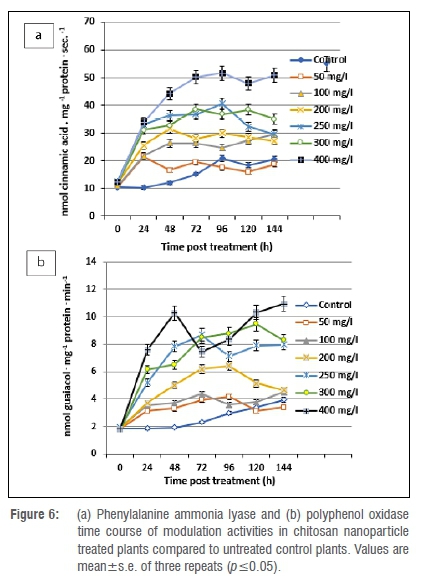
Modulations in enzyme activity
Phenylalanine ammonia lyase activity was considerably increased by 1.00-fold in all ChiNP-treated plants at 24 h post spraying. The maximum increase in phenylalanine ammonia lyase activity was noted in 400 mg/L treated plants. The peak activity continually increased to reach its maximum level at 144 h with a 3.14-fold increase, while the activity increased only 0.93-fold in untreated control plants (Figure 6a). The maximum polyphenol oxidase activity was recorded in plants treated with 400, 300 and 250 mg/L ChiNPs in which the increase in polyphenol oxidase activity was found to be considerably higher than in untreated controls. However, a slight increase in polyphenol oxidase activity was generated after the virus treatment, with a 0.28-fold increase at 72 h post-treatment, reaching a maximum of 1.11-fold at 144 h (Figure 6b).
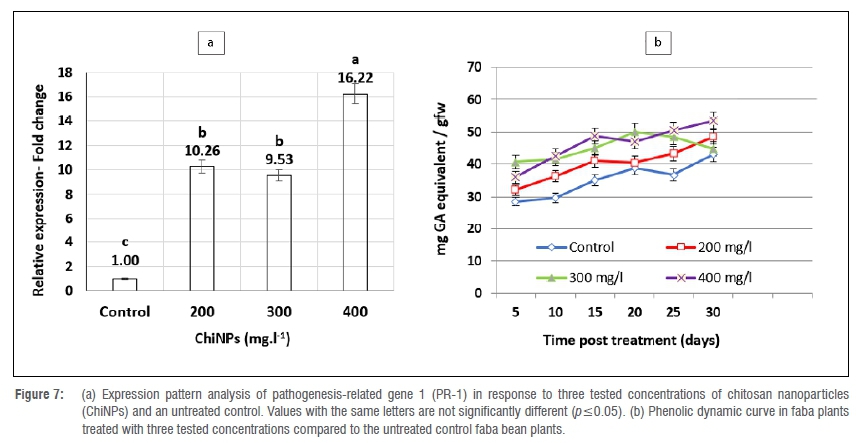
Changes in gene expression level
ChiNP foliar spraying was found to modulate the plant defence machinery by upregulating the PR-1 gene (Figure 7a). The gene transcriptome was strongly promoted with all tested applications of ChiNPs compared to untreated control plants. The relative expression increased as the ChiNP concentration increased. The maximum upregulating expression level of a 16.22-fold change was attained with 400 mg/L. Simultaneously, the mRNA accumulation of the PR-1 gene in 200- and 300 mg/L treated plants was high, with 10.26- and 9.56-relative expression fold increases, respectively, over the untreated control plants.
Changes in total phenolic content
Total phenolic content was relatively affected in faba bean plants treated with the tested ChiNP concentrations (Figure 7b). Among all tested concentrations of ChiNPs, only the 400 mg/L dosage rate increased the total phenolic content curve significantly over untreated controls until 30 days post-application, while 300 mg/L significantly increased the total phenolic content for 20 days compared to controls.
Effects of ChiNPs on plant vegetation and growth parameters
Foliar application of ChiNPs significantly affected plant growth, leaf area and chlorophyll content parameters compared to untreated infected controls. Moreover, 400 mg/L ChiNPs not only produced a significant increase in shoot length in comparison to the untreated infected controls, but also increased plant growth more than that of the untreated healthy control plants (Table 2).
Discussion
The nanotechnology approach has recently emerged as a new potential means of control for a wide array of plant diseases.35 However, the use of nanotechnology in plant protection management has not been broadly introduced on large scales as yet, and is still under research at a small scale of production and application.36,37 The current study shows the antiviral activity of ChiNPs against BYMV and its accumulation within the plant tissues as compared to the bulk form of chitosan. To our knowledge, this is the first comprehensive study to investigate the antiviral ability of ChiNPs against plant viruses. However, previous reports have confirmed that ChiNPs increased the antiviral capabilities against human viruses such as hepatitis C virus38 and HIV39. Furthermore, very limited reports have also shown the partial effectiveness of bulk chitosan polymers against plant viruses.40
The mechanisms underlying the effectiveness of ChiNPs in controlling plant viruses and other plant diseases have not as yet been investigated. Nanoparticles, ranging from 1 nm to 100 nm in size, provide superior chemical and physical features when compared to their bulk materials due to their large surface area to volume ratios.910 Because all plant viruses are biological nano-size particles, it is reasonable to argue that chitosan in nano-based form could facilitate antiviral activity by targeting the virus particles and the replication process within the host tissues. Further, our findings show that ChiNPs altered the virus integrity by producing incomplete and defective viral particles. A potential reason for this may be that the synthesised ChiNPs have a surface area with positive charges and positively charged amino gatherings on chitosan glucosamine polymer chains. These facts support the suggestion that ChiNPs might have high bio-reactivity to attract viral RNA which contains negatively charged phosphate groups in its primary chain,4142 and thereby suppresses virus replication and disease progression. Furthermore, as all viral proteins have negatively charged clusters of glycol proteins, it is assumed that the positively charged nanoparticles could also target the virus coat protein.43 This supports our suggestion that the role of ChiNPs in controlling the plant viruses might be strongly governed by their nano physicochemical properties, particle chemical nature and bio-reactivity, in which the chitosan in nano-size seems to be critical for its antiviral properties.
In parallel with these findings, our research also suggests that the ChiNPs promote the plant defence mechanism against virus invasion by promoting expression of the PR-1 gene and some defence-related enzymes (phenylalanine ammonia lyase, polyphenol oxidase) and increasing the total phenolic content. The initiation of plant defence boosters in treated plants may be attributable to the ability of chitosan to modulate salicylic acid phytohormone pathways which include the synthesis of phenylalanine ammonia lyase and pathogenesis-related proteins.44 This supports the results of Chandra et al.20 who investigated the role of ChiNPs as a natural biopolymer in the immunomodulatory response. Chitosan was found to enhance anti-oxidising enzymes and total phenolic content that are involved in the plant defence response. Moreover, Jia et al.45 found that chitosan oligosaccharide spraying boosted the innate immunity system in treated plants by upregulating the PR-1 defence-related gene and increased the level of salicylic acid in treated plants. Further, Jia et al.45 investigated how to reduce the virus accumulation content and symptom development by activating plant hormone signalling pathways. The increase in total phenolic contents may strongly be associated with increasing phenylalanine ammonia lyase activity, which is the first key enzyme for producing phenolics and other secondary metabolites involved in the plant defence system against pathogen attacks.46
Our data suggest that chitosan nano applications have a dual role in controlling plant viral infection and disease expression. Our data demonstrate that ChiNP application prevented the stunting of the plant usually caused by the virus, by reducing the virus infectivity, and thereby increasing growth parameters even above those of the non-infected untreated controls. The role of the chitosan polymer in promoting plant growth and yield has been documented in numerous crops.47-50 Furthermore, chitosan nanoparticles have been investigated as plant growth enhancers.51 The mode of action underlying the role of chitosan in enhancing plant growth was proposed to be its ability to induce many physiological processes including nutrient uptake, photosynthesis, and cell division as well as plant hormones.52 Thus, it is reasonable to assume that chitosan in its nano-based form could be curatively used to alleviate viral infections.
Conclusion
In conclusion, the current findings provide an indication of the potency of chitosan nano-based materials against plant viruses. ChiNPs showed a powerful antiviral activity against BYMV infectivity, virus replication and accumulation in treated plants. This might offer a new alternative strategy to manage plant viral diseases without the use of pesticides. However, further research to support this initial study is needed to explore the mechanisms underlying the role of ChiNPs against plant viruses.
Acknowledgements
We thank Dr Khaled Y. Farroh, Head of the Central Laboratory of Nanotechnology & Advanced Materials, Agricultural Research Center (Giza, Egypt), for providing laboratory facilities for the characterisation of chitosan nanoparticles. We also thank Dr Yosra Ahmed, Head of the Plant Quarantine Pathogens Laboratory, Plant Pathology Research Institute, Agricultural Research Center (Giza, Egypt) for providing chemical reagents and consumables for the PR-1 gene expression experiment.
Competing interests
We have no competing interests to declare.
Authors' contributions
A.Y.E.: Conceptualisation, methodology, data collection, data analysis, writing - the initial draft. M.M.A.: Validation, supervision, writing -revisions. T.E.: Methodology, data analysis. M.I.A.: Supervision, review and editing. M.R.T.: Supervision, writing - revisions. All authors revised the initial draft of the manuscript and approved the final draft.
References
1. Bos L. Hundred years of virology: From vitalism via molecular biology to genetic engineering. Trends Microbiol. 2000;8:82-104. https://doi.org/10.1016/S0966-842X(99)01678-9 [ Links ]
2. Singh AK, Bharati RC, Manibhushan NC, Pedpati A. An assessment of faba bean (Vicia faba L.) current status and future prospects. Afr J Agric Res. 2013;8(50):6634-6641. https://academicjournals.org/article/article1387443711_Singh%20et%20al.pdf [ Links ]
3. Sharma PN, Sharma V Anuradha S, Rajput K, Sharma SK. Identification and molecular characterization of Bean yellow mosaic virus infecting French bean in Himachal Pradesh. Virus Diseases. 2015;26(4):315-318. https://doi.org/10.1007/s13337-015-0270-z [ Links ]
4. Khalil SA, Erskine W. Combating disease problems of grain legumes in Egypt. Grain Legumes. 2001;32:24-26. [ Links ]
5. Latham LJ, Jones RA. Incidence of virus infection in experimental plots, commercial plots, and seed stocks of cool-season crop legumes. Aust J Agric Res. 2001;52:397-413. https://doi.org/10.1071/AR00079 [ Links ]
6. Hampton RO, Jensen A, Hagel GT. Attributes of Bean yellow mosaic potyvirus transmission from clover to snap beans by four species of aphids (Homoptera; Aphididae). J Econ Entomol. 2005;98(6):1816-1823. https://doi.org/10.1603/0022-0493-98.6.1816 [ Links ]
7. Jones RA. Trends in plant virus epidemiology: Opportunities from new improved technologies. Virus Res. 2014;186:3-19. https://doi.org/10.1016/j.virusres.2013.11.003 [ Links ]
8. Miller G. Sustaining the earth. 6th edition. Pacific Grove, CA: Thompson Learning; 2004. [ Links ]
9. Osuwa JC, Anusionwu PC. Some advances and prospects in nanotechnology: A review. Asian J Inform Technol. 2011 ;10(2):96-100. https://doi.org/10.3923/ajit.2011.96.100 [ Links ]
10. Bakshi M, Singh HB, Abhilash PC. The unseen impact of nanoparticles: More or less? Current Sci. 2014;106(3):350-352. [ Links ]
11. Hadwiger LA, Beckman JM. Chitosan as a component of pea-Fusarium solani interactions. J Plant Physiol. 1980;66:205-211. https://doi.org/10.1104/pp.66.2.205 [ Links ]
12. Wu T, Zivanovic S, Draughon FA, Conway WS, Sams CE. Physicochemical properties and bioactivity of fungal chitin and chitosan. Agric Food Chem. 2005;53:3888-3894. https://doi.org/10.1021/jf048202s [ Links ]
13. Xing K, Zhu X, Peng X, Qin S. Chitosan antimicrobial and eliciting properties for pest control in agriculture. Rev Agron Sustain Dev. 2015;35:569-588. https://doi.org/10.1007/s13593-014-0252-3 [ Links ]
14. Chirkov S. The antiviral activity of chitosan (review). Appl Biochem Microbiol. 2002;38:1-8. https://doi.org/10.1023/A:1013206517442 [ Links ]
15. Chirkov S, Il'ina A, Surgucheva N, Letunova E, Varitsev Y Tatarinova N, et al. Effect of chitosan on systemic viral infection and some defense responses in potato plants. Russian J Plant Physiol. 2001;48:774-779. https://doi.org/10.1023/A:1012508625017 [ Links ]
16. Atia MM, Buchenauer H, Aly AZ, Abou-Zaid MI. Antifungal activity of chitosan against Phytophthora infestans and activation of defence mechanisms in tomato to late blight. Biol Agric Hortic. 2005/23(2):175-197. https://doi.org/10.1080/01448765.2005.9755319 [ Links ]
17. El Gamal AYS, Zayed MA, El-Nashar FK, Atia MM. The potential ability of some abiotic agents to control barley net blotch disease. Zagazig J Agric Res. 2016;43(4):1215-1232. https://doi.org/10.21608/zjar.2016.100490 [ Links ]
18. Mishra S, Jagadeesh KS, Krishnaraj PU, Prem S. Biocontrol of tomato leaf curl virus (TLCV) in tomato with chitosan supplemented formulations of Pseudomonas sp. under field conditions. Aus J Crop Sci. 2017;8:347-355. [ Links ]
19. Firmansyah D, Hidayat S, Widodo W. Chitosan and plant growth-promoting rhizobacteria application to control squash mosaic virus on cucumber plants. Asian J Plant Pathol. 2017;11:148-155. https://doi.org/10.3923/ajppaj.2017.148.155 [ Links ]
20. Chandra S, Chakraborty N, Dasgupta A, Sarkar J, Panda K, Acharya K. Chitosan nanoparticles: A positive modulator of innate immune responses in plants. Sci Rep. 2015;5:1-13. https://doi.org/10.1038/srep15195 [ Links ]
21. Clark MF, Adams AN. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol. 1977;34:475-483. https://doi.org/10.1099/0022-1317-34-3-475 [ Links ]
22. Kuhn CW. Separation of cowpea virus mixtures. Phytopathology. 1964;54:739-740. https://doi.org/10.1016/jjviromet.2008.07.023 [ Links ]
23. 23 AL-Khalifa M, Kumari SG, Kasem A, Makkouk KM, Shalaby AA, AL-Chaabi SA. Molecular characterization of a Bean yellow mosaic virus isolate from Syria. Phytopathol Mediterr. 2008;47:282-285. https://doi.org/10.14601/Phytopathol_Mediterr-2734 [ Links ]
24. Masarudin MJ, Cutts SM, Evison BJ, Phillips DR, Pigram PJ. Factors determining the stability, size distribution, and cellular accumulation of small, monodisperse chitosan nanoparticles as candidate vectors for anticancer drug delivery: Application to the passive encapsulation of [14 C]-doxorubicin. Nanotechnol Sci Appl. 2015;8:67-80. https://doi.org/10.2147/NSA.S91785 [ Links ]
25. Zhang H, Wu F, Li YYang X, Huang J, Lv T, et al. Chitosan-based nanoparticles for improved anticancer efficacy and bioavailability of mifepristone. Beilstein J Nanotechnol. 2016;7:1861-1870. https://doi.org/10.3762/bjnano.7178 [ Links ]
26. Yang X, Liangyi K, Tien P. Resistance of tomato infected with cucumber mosaic virus satellite RNA to potato spindle tuber viroid. Ann Appl Biol. 1996;129:543-551. https://doi.org/10.1111/j.1744-7348.1996.tb05775.x [ Links ]
27. Campos RE, Bejerman N, Nome C, Laguna IG, Pardina PR. Bean Yellow Mosaic Virus in Soybean from Argentina. J Phytopathol. 2013;162:322-325. https://doi.org/10.1111/jph.12185 [ Links ]
28. Kar M, Mishra D. Catalase, peroxidase, and polyphenol oxidase activities during rice leaf senescence. Plant Physiol. 1976;57:315-319. https://doi.org/10.1104/pp.57.2.315 [ Links ]
29. Lisker N, Coren L, Chalutz E, Fucus Y Fungal infections suppress ethylene-induced phenylalanine ammonia-lyase activity in grape fruits. Physiol Plant Pathol.1983;22:331-338. https://doi.org/10.1016/S0048-4059(83)81020-0 [ Links ]
30. Cheng Y Zhang H, Yao J, Wang X, Xu J, Han Q, et al. Characterization of non-host resistance in broad bean to the wheat stripe rust pathogen. BMC Plant Biol. 2012;12:96. https://doi.org/10.1186/1471-2229-12-96 [ Links ]
31. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001 ;29(9):1-45. https://doi.org/10.1093/nar/29.9.e45 [ Links ]
32. Lin JY Tang CY Determination of total phenolic and flavonoid contents in selected fruits and vegetables, as well as their stimulatory effects on mouse splenocyte proliferation. Food Chem. 2007;101 (1 ):140-147. https://doi.org/10.1016/j.foodchem.2006.01.014 [ Links ]
33. Chahit D. A leaf area estimation model for faba bean (Vicia faba L.) grown in the Mediterranean type of climate. Agric J SDU. 2012;7(1):58-63. [ Links ]
34. Siliva FDA, Azevedo CA. Principal components analysis in the software Assistat-statistical attendance. In: World Congress on Computers in Agriculture. Reno, NV: American Society of Agricultural and Biological Engineers; 2009. https://www.scirp.org/(S(i43dyn45teexjx455qlt3d2q) [ Links ]
35. Singh S, Singh BK, Ydava SM, Kupta AK. Applications of nanotechnology in agriculture and their role in disease management. Res J Nanosci Nanotechnol. 2015;5(1):1-5. https://doi.org/10.3923/rjnn.2015.L5 [ Links ]
36. Prasad R, Bhattacharyya A, Nguyen QD. Nanotechnology in sustainable agriculture: Recent developments, challenges, and perspectives. Front Microbiol. 2017;8:1014. https://doi.org/10.3389/fmicb.2017.01014 [ Links ]
37. Luca M. Nanotechnology in agriculture: New opportunities and perspectives. New Visions in Plant Science. IntechOpen; 2018. https://doi.org/10.5772/intechopen.74425 [ Links ]
38. Loutfy S, Elberry M, Farroh K, Mohamed H, Mohamed A, Mohamed E, et al. Antiviral activity of chitosan nanoparticles encapsulating curcumin against Hepatitis C Virus genotype 4a in human hepatoma cell lines. Int J Nanomed. 2020;15:2699-2715. https://doi.org/10.2147/IJN.S241702 [ Links ]
39. Ramana LN, Sharma S, Sethuraman S, Ranga U, Krishnan UM. Evaluation of chitosan nanoformulations as potent anti-HIV therapeutic systems. Biochim Biophys Acta. 2014;1840(1 ):476-484. https://doi.org/10.1016/j.bbagen.2013.10.002 [ Links ]
40. Iriti M, Sironi M, Gomarasca S, Casazza AP Soave C, Faoro F. Cell death-mediated antiviral effect of chitosan in tobacco. Plant Physiol Biochem. 2006;44:893-900. https://doi.org/10.1016/j.plaphy.2006.10.009 [ Links ]
41. Xing K, Chen XG, Liu CS, Cha DS, Park HJ. Oleoyl-chitosan nanoparticles inhibit Escherichia coli and Staphylococcus aureus by damaging the cell membrane and putative binding to extracellular or intracellular targets. Int J Food Microbiol. 2009;132:127-133. https://doi.org/10.1016/j.ijfoodmicro.2009.04.013 [ Links ]
42. Mansilla AY Albertengo L, Rodriguez MS, Debbaudt A, Zúniga A, Casalongué CA. Evidence on antimicrobial properties and mode of action of a chitosan obtained from crustacean exoskeletons on Pseudomonas syringae pv. tomato DC3000. Appl Microbiol Biotechnol. 2013;97:6957-6966. https://doi.org/10.1007/s00253-013-4993-8 [ Links ]
43. Karlin S, Brendel V Charge configurations in viral proteins. Proc Natl Acad Sci USA. 1988;85(24):9396-9400. https://doi.org/10.1073/pnas.85.24.9396 [ Links ]
44. Siddaiah S, Prashanth H, Satyanarayana N, Mudili V Gupta V Kalagatur N, et al. Chitosan nanoparticles having higher degree of acetylation induce resistance against pearl millet downy mildew through nitric oxide generation. Sci Rep. 2019;8:101-116. https://doi.org/10.1038/s41598-017-19016-z [ Links ]
45. Jia X, Meng Q, Zeng H, Wang W, Yin H. Chitosan oligosaccharide induces resistance to Tobacco mosaic virus in Arabidopsis via the salicylic acid-mediated signaling pathway. Sci Rep. 2016;6:26144. https://doi.org/10.1038/srep26144 [ Links ]
46. Dogbo DO, Gogbeu SJ, Zuel B, Yao NK, Zohouri GP Mamyrbekov-Abekro JA, et al. Comparative activities of phenylalanine ammonia-lyase and tyrosine ammonia-lyase and phenolic compounds accumulated in cassava elicited cell. Afr Crop Sci. 2012;20(2):85-94. [ Links ]
47. Lee YS, Kim YH, Kim SB. Changes in the respiration, growth, and vitamin C content of soybean sprouts in response to chitosan of different molecular weights. Hortic Sci. 2005;40:1333-1335. https://doi.org/10.21273/HORTSCI.40.5.1333 [ Links ]
48. Mondal MMA, Malek MA, Puteh AB, Ismail MR, Ashrafuzzaman M, Naher L. Effect of foliar application of chitosan on growth and yield in okra. Aust J Crop Sci. 2012;6(64):918-921. [ Links ]
49. Sultana S, Islam M, Khatun MA, Hassain MA, Huque R. Effect of foliar application of oligo-chitosan on growth, yield and quality of tomato and eggplant. Asian J Agric Res. 2017;11:36-42. https://doi.org/10.3923/ajar.2017.36.42 [ Links ]
50. Akter J, Jannat R, Hossain MM, Ahmed JU, Rubayet MT. Chitosan for plant growth promotion and disease suppression against anthracnose in chili. Int J Agric Environ Biotechnol. 2018;3:806-817. https://doi.org/10.22161/ijeab/3.3.13 [ Links ]
51. Choudhary RC, Kumaraswamy RV Kumari S, Sharma SS, Pal A, Raliya R, et al. Cu-chitosan nanoparticle boost defense responses and plant growth in maize (Zeamays L.). Sci Rep. 2017;7:9754-9765. https://doi.org/10.1038/s41598-017-08571-0 [ Links ]
52. Chakraborty M, Hasanuzzaman M, Rahman M, Khan M, Bhowmik P Mahmud N, et al. Mechanism of plant growth promotion and disease suppression by chitosan biopolymer. Agriculture. 2020;10:624. https://doi.org/10.3390/agriculture10120624 [ Links ]
 Correspondence:
Correspondence:
Ahmed El Gamal
Email: ahmedvnp1@yahoo.com
Received: 09 Apr. 2021
Revised: 21 July 2021
Accepted: 28 July 2021
Published: 27 Jan. 2022
FUNDING: None
EDITOR: Teresa Coutinho














