Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.117 no.11-12 Pretoria nov./dic. 2021
http://dx.doi.org/10.17159/sajs.2021/8667
RESEARCH ARTICLE
Antibiotic resistance profiles of Staphylococcus spp. from white button mushrooms and handlers
Stacey Duvenage; Werner Rossouw; Germán Villamizar-Rodríguez; Erika M. du Plessis; Lise Korsten
DSI-NRF Centre of Excellence in Food Security, Department of Plant and Soil Sciences, University of Pretoria
ABSTRACT
The presence of Staphylococcus spp. has increasingly been reported in food products and poses a public health threat. The aim of this study was to determine the diversity of Staphylococcus spp. and the antibiotic resistance profiles of isolates obtained from freshly harvested and packed ready-to-eat mushrooms (n=432) and handlers' hands (n=150). A total of 56 Staphylococcus isolates [46.4% (n=26) from hands and 53.6% (n=30) from mushrooms] were recovered belonging to 10 species. Staphylococcus succinus isolates (n=21) were the most prevalent, of which 52.4% came from mushrooms and 47.6% from hands. This was followed by S. equorum isolates [n=12; 91.7% (n=11) from mushrooms and 8.3% (n=1) from hands] and S. saprophyticus [n=9; 66.7% (n=6) from mushrooms and 33.3% (n=3) from hands]. Six isolates that were characterised as multidrug resistant were isolated from hands of handlers. Most (83.9%; n=47) of the 56 isolates were resistant to penicillin [53.2% (n=25) from mushrooms and 46.8% (n=22) from hands] and 14.3% (n=8) were resistant to cephalosporin classes [25% (/=2) from mushrooms and 75% (n=6) from hands], both of which are used to treat staphylococcal infections. Antibiotic resistance genes blaZ [25.0% (n=14) of all isolates of which 71.4% (n=10) were from hands and 28.57% (n=4) from mushrooms], tetL and tetK [both 1.8% (n=1) from hands], mecA [5.4% (n=3) from hands] and ermA [1.8% (n=1) from mushrooms] were detected from the 56 isolates. Only two (25.0%) of the eight methicillin-resistant staphylococci harboured the mecA gene, while only 11 (23%) of the 47 penicillin-resistant isolates harboured the blaZ gene [36.4% (n=4) from mushrooms and 63.6% (n=7) from hands]. Our results demonstrate that food handlers and harvested and packed ready-to-eat mushrooms could be a source of diverse Staphylococcus spp. that exhibit antimicrobial resistance. Clinically relevant S. aureus was only detected on one handler's hand; however, the isolate was not multidrug resistant. The presence of diverse Staphylococcus spp. on mushrooms and the hands of handlers is a potential public health concern due to their potential to cause opportunistic infections.
SIGNIFICANCE:
• This study is the first to describe the antibiotic resistance profiles and antibiotic gene presence of Staphylococcus spp. isolated from fresh mushrooms and hands of pickers and packers. Mushrooms and handlers in this study were demonstrated to be possible routes of transmission of Staphylococcus spp. that are antibiotic resistant and which harbour antibiotic resistance genes, presenting a possible public health hazard.
Keywords: staphylococci, antibiotic genes, antibiotic resistance
Introduction
Staphylococcus spp. are ubiquitous and transient organisms. Staphylococcus aureus is known to be a natural coloniser of human skin, and between 10% and 20% of adults' skin is persistently colonised by S. aureus, while between 30% and 50% of healthy people's skin is colonised by S. aureus.1,2 Staphylococcus aureus specifically has been recorded to cause invasive infections or toxin-mediated diseases, including endocarditis, bacteraemia, metastatic infections and toxic shock syndrome.3 Traditionally, much attention has been paid to S. aureus as an organism causing infection; however, coagulase-negative Staphylococcus spp. have more recently been shown to also be pathogenic.4
Treatment of staphylococcal infections with antibiotics has become common practice in the medical field and, subsequently, higher levels of resistance have been recorded.2,5 The spread of antimicrobial-resistant staphylococci represents a hazard to human and veterinary health6 because staphylococci have the ability to transfer antibiotic resistance genes (ARGs) to other pathogenic organisms2,5. Previous studies have also shown that staphylococci can be important reservoirs of ARGs in ready-to-eat food.7,8 Moreover, the exchange of genetic material, such as mobile elements, has reportedly been associated with food-processing environments. 9
Multidrug-resistant (MDR) staphylococci add to the public health concerns with respect to staphylococcal infections. For example, it has been associated with an increased severity of infections as well as a growing number of people being hospitalised due to such infections.10 In the United States of America, 60% of infectious disease specialists reported that untreatable bacterial infections had been observed, highlighting the impact on human health.11 The prevalence of MDR organisms in environmental samples has further heightened the concern of staphylococcal infections for the World Health Organization10 due to the potential horizontal transfer of ARGs, genetic mutation and recombination. Food associated with bacteria harbouring such ARGs is a major concern and represents possible reservoirs for the spread of these ARGs.6
Food commodities that are handled extensively and do not go through a decontamination step might therefore harbour antimicrobial-resistant microorganisms and thus pose a human and environmental health threat. Food products harbouring antimicrobial-resistant Escherichia coli, Campylobacter spp., Salmonella spp., Clostridium spp. and Listeria monocytogenes have previously been documented and there has now been an increased interest in the role of Staphylococcus spp. isolated from food.2,6 Therefore, the aim of this study was to determine the diversity of Staphylococcus spp. as well as the antibiotic resistance (AR) profiles and the presence of ARGs in isolates obtained from mushrooms and mushroom handlers. Mushrooms that are handled extensively by pickers and packers represent a potential risk if hygiene principles are not observed. This study is the first of its kind to investigate the diversity and AR profiles of Staphylococcus spp. on mushrooms and handlers' hands.
Materials and methods
A total of 432 white button mushroom samples were collected as outlined in Rossouw and Korsten12. Sampling sites included two large-scale commercial mushroom farms located in Gauteng Province, South Africa, which follow similar production practices, operating under Global-G.A.P Integrated Farm Assurance Standards V5.1. Mushrooms that were void of defects and at the ready-to-harvest stage were randomly sampled based on uniformity of size, shape and maturity. Mushrooms were aseptically harvested by researchers.12 Packed mushrooms handled by pickers and packers prior to punnet sealing were also collected. Sampling of hands was done according to standard hand swab procedures.13 Hand swabs were collected using the Copan Venturi Transystem (Copan, Italy) from five pickers' and five packers' dominant hands on a weekly basis for a period of 15 weeks, resulting in 150 hand samples. Samples were placed in a cooler box and transported to the laboratory for analysis within 24 h. At the time of the study, ethical clearance for non-invasive swabbing of the hands was not mandatory and was therefore waived by the Ethics Committee of the Faculty of Natural and Agricultural Sciences, University of Pretoria. Commercial and personal permission to conduct non-invasive hand swabbing was granted by the commercial entity as well as by the persons whose hands were sampled. Confidentiality was maintained throughout the process.
A sample (250 g) was aseptically obtained from each mushroom sample and homogenised using a handheld blender (Russell Hobbs, Johannesburg, South Africa). Ten grams of the homogenised sample was added to 90 mL tryptone soy broth (Merck, Johannesburg) in a sterile homogeniser bag, macerated for 5 min and incubated for 24 h at 37 °C. Contents were subsequently plated onto Baird-Parker agar (Merck) and incubated for 24 h at 37 °C.
Hand swabs were placed into 9 mL buffered peptone water (Merck), and incubated for 24 h at 37 °C and plated onto Baird-Parker agar plates, which were incubated for 24 h at 37 °C. Presumptive Staphylococcus spp. were selected from the Baird-Parker agar, based on a dark grey to black colony morphology which was surrounded by a clear zone.14 Isolates were purified on Baird-Parker agar and the purified isolate identity was determined using matrix-assisted laser desorption ionisation time-of-flight mass spectrometry in combination with the Bruker Biotyper software and database.15
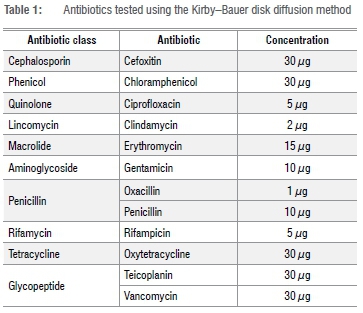
A total of 56 purified and confirmed Staphylococcus spp. isolates were used for further characterisation of AR, using the Kirby-Bauer disk diffusion method.16 Each isolate was cultured in 9 mL of brain heart infusion broth (Merck) and incubated for 18 h at 37 °C (which resulted in approximately log 8 cfu/mL17) and subsequently plated onto Mueller- Hinton agar plates (Merck). The Kirby-Bauer disk diffusion test was employed to determine susceptibility to the antibiotics listed in Table 1. Zone diameters were measured and interpreted according to the Clinical and Laboratory Standards Institute guidelines.17 Strains resistant to three or more antibiotic classes were defined as MDR.
Genomic DNA (gDNA) was extracted using the Quick-gDNA Miniprep kit (Zymo Research, Irvine, CA, USA), following overnight incubation in tryptone soy broth at 37 °C. The DNA concentration was measured using the broad range kit for Qubit 2.0 fluorometer (Life Technologies, Johannesburg, South Africa). Polymerase chain reaction (PCR) mixtures with a final volume of 25 μL, containing 10-100 μg gDNA, were prepared using Dream Taq PCR Master Mix (1x) (Thermo Scientific, Johannesburg, South Africa). Each reaction included 16S universal primers as amplification controls (Table 2) as well as the specific ARG primer with primer concentration as outlined in Table 2. PCR cycling conditions were as follows: 94 °C for 2 min, followed by 35 cycles of denaturation (30 s at 95 °C), annealing (30 s at primer temperature, Table 1) and extension (60 s at 72 °C), with a final extension at 72 °C for 5 min. PCR reactions were performed on a BioRad T100 thermocycler (BioRad, Johannesburg, South Africa). Amplicons were visualised on a 2% agarose gel stained with Roti®-safe (Carl Roth GmbH & Co, Karlsruhe, Germany) using a molecular imager in conjunction with the Image Lab™ software (BioRad).
Results
Out of 582 samples collected, 56 (9.6%) were positive for Staphylococcus spp. All Staphylococcus spp. isolates and their AR and ARG profiles are presented in Figure 1 by sample and year collected. Ten Staphylococcus species were isolated: S. aureus, S. epidermidis, S. equorum, S. haemolyticus, S. hominis, S. saprophyticus, S. sciuri, S. succinus, S. warneri and S. xylosus (Figures 1 and 2). Staphylococcus aureus was only isolated from one packer's hand sample and no mushroom sample yielded S. aureus. Staphylococcus epidermidis was detected from one picker's and one packer's hand, and both isolates were found to be MDR. A total of 12 S. equorum isolates were detected and were isolated mainly from hands (91.67%; n=11/12) of pickers (41.67%; n=5/12) and packers (50%; n=6/12), with only 8.33% (n=1/12) isolated from a packed mushroom sample. Two MDR S. equorum isolates from pickers' hands, isolated from the same farm and same mushroom picking room (data not shown), shared a phenotypic MDR antibiotic profile but the ARGs were different. Only one MDR S. haemolyticus was isolated, from a packer's hand. One packer's hand and one packed mushroom sample were contaminated with S. hominis, with the one on the packed mushrooms found to be MDR. Mushrooms sampled before harvest (n=6) and three pickers' hands were contaminated with S. saprophyticus. Only one S. sciuri was isolated from one packed mushroom sample. Staphylococcus succinus was isolated from hands (n=10) as well as from mushrooms before picking (n=4) and after packing (n=7), with one found to be methicillin resistant due to the presence of mecA. Four of the six S. succinus isolates were isolated from pickers in the same growing room during the same sampling period (data not shown). Staphylococcus xylosus was only isolated from mushrooms sampled before picking (n=4) and after packing (n=2) - these isolates were resistant to only the penicillin class of antibiotics. Staphylococcus warneri was isolated from a packer's hand.
Six Staphylococcus spp. (10.7%; n=6/56) were considered MDR, with all isolated from handlers' hands. Expressed resistance to at least one antibiotic agent was found in 85.7% [n=48; 52.1% (n=25) from mushrooms and 47.9% (n=23) from hands] of all Staphylococcus spp. characterised. A total of 83.9% of the 56 isolates [n=47; 53.2% (n=25) from mushrooms and 46.8% (n=22) from hands] were resistant to the penicillin class. Resistance to both penicillin and oxacillin was observed in 32 of the 56 isolates, of which 46.9% (n=15) were from mushrooms and 53.1% (n=17) were from hands.
Of the 47 isolates resistant to the penicillin class, 23.4% (n=11) harboured the blaZ gene and 6.4% (n=3) harboured the blaZ gene but did not express resistance to penicillin nor oxacillin. Of the 11 that harboured the blaZ gene, 36.4% (n=4) were from mushrooms and 63.6% (n=7) were from hands. Of the 32 isolates resistant to oxacillin, 9.4% (n=3) of isolates from hands harboured the mecA gene. Three isolates from hands were resistant to oxytetracycline, with one isolate from hands harbouring tetK and one isolate from hands harbouring tetL. Only one mushroom isolate harboured ermA. No ampC was detected, even though eight Staphylococcus spp. were resistant to cefoxitin. Two of the three isolates from hands harbouring mecA were resistant to cefoxitin.
Discussion
The status of antibiotic-resistant microorganisms in the agricultural environment is becoming a major concern in the global food industry and in the public health sector.5 The prevalence and status of staphylococci in animal products like meat and cheese have been well established.5,8,14,18-20However, there is limited information on the prevalence of AR and ARG in Staphylococcus spp. isolated from fresh produce. This study, the first of its kind as far as we could determine, investigated the diversity and resistance of Staphylococcus spp. isolated from white button mushrooms and mushroom handlers.
Of the ten Staphylococcus species identified, seven were found to be associated with raw mushrooms - these were S. epidermidis, S. equorum, S. hominis, S. saprophyticus, S. sciuri, S. succinus and S. xylosus. Both S. aureus and S. epidermidis are associated with the human skin microflora; these as well as other staphylococci pose an important health concern, in addition to exhibiting AR. The presence of these species associated mainly with the hands of workers indicates the need for improved personal hygiene implementation, training and enforcement.2 These isolates can contribute to illness of both the handlers as well as susceptible consumers.
In this study, S. saprophyticus, S. succinus and S. xylosus were mainly detected from mushrooms. A previous study determined the presence of S. xylosus, S. epidermidis and S. saprophyticus in ready-to-eat products from animal origin. Furthermore, these species are also commonly associated with farm animals.8 Mushrooms are cultivated on compost composed of hay, chicken manure, leachate and agricultural lime, which are stored on farms for months in bulk, prior to composting. Antibiotic-resistant bacteria have been found to be present in the excreta of broiler chickens.21 Graham et al.21 concluded that typical storage of chicken manure was not sufficient to eliminate the antibiotic-resistant Staphylococcus spp. Compost used for the production of mushrooms, which includes chicken manure, is pasteurised at between 60 °C and 75 °C for 13 days. However, Fontes et al.19 found that staphylococci were able to survive high temperature pasteurisation. Therefore, the presence of these species is not necessarily eliminated from mushroom compost during the pasteurisation process. In addition, previous research has demonstrated that the use of antibiotics in the chicken rearing industry has been linked to the presence of antibiotic-resistant organisms present on farm workers as well as the growing environment.23 Moreover, flies have previously been reported to increase the human exposure to antibiotic-resistant bacteria.23 The presence of Staphylococcus spp. could be due to survival of organisms during storage and pasteurisation because of the organism's innate ability to survive such conditions as well as the possibility of post-pasteurisation contamination. All these factors might lead to the establishment and spread of Staphylococcus species, and antibiotic-resistant and MDR staphylococci in the environment and food system.
There has been an increase in the number of antibiotic-resistant organisms associated with humans2, their direct environments20,22,24 and their food2,8,14,18,19,25. Transmission of AR in bacteria is further aided by the ability of food to be a vehicle.8 In the current study, only 10% of all staphylococci isolates were considered MDR. Benjelloun Touimi et al.2 found that 100% of Staphylococcus isolates from vegetable and food handlers were MDR. Moreover, in ready-to-eat food, 94.12%25, 89.81%22 and 32.8%8 of Staphylococcus isolates investigated were found to be MDR, reaffirming that food can be a vehicle for the spread of MDR Staphylococcus spp.
Penicillin, oxacillin and methicillin are the first line of defence against clinical staphylococci infections. In this study, 83.9% of isolates were resistant to the penicillin class, with 14 isolates harbouring blaZ, the gene that encodes β-lactamase and confers resistance to penicillin. Similarly, 94% of Staphylococcus spp. isolated from chicken and beef14, 78.5% from soft cheeses19 and 67.4% from cow mastitis20 were resistant to penicillin. Moreover, Benjelloun Touimi et al.2 found that all Staphylococcus spp. isolates from ready-to-eat foods were resistant to penicillin and oxacillin. Klimiene et al.20, however, found a 66% presence of the blaZ gene in staphylococci assessed, compared with our study's 25% blaZ gene prevalence.
The resistance of strains to cefoxitin is an indication of methicillin resistance in Staphylococcus phenotypes which are resistant to all β-lactam antibiotics, including penicillin and isoxazolyl penicillins.8 The mecA gene confers resistance to methicillin (oxacillin and /or cefoxitin) and its potential transfer between organisms represents a public health concern.2 In this study, 14.3% (n=8/56) of isolates were resistant to cefoxitin, of which 25.0% (n=2/8) were found on mushrooms and 75.0% (n=6/8) were found on hands; however, only two of the cefoxitin-resistant isolates harboured the mecA gene. Vyletelova et al.26VLKOVAH.,MANGA I. (2011 found that not all isolates (174/200) that showed resistance to cefoxitin harboured mecA. Chajçcka-Wierzchowska et al.8 found that all but one isolate exhibiting cefoxitin resistance harboured mecA, which is part of the staphylococcal cassette chromosome mec (SCCmec) on the bacterial chromosome. All isolates in the current study that were resistant to cefoxitin were also resistant to oxacillin and penicillin. The co-occurrence of resistance to methicillin, oxacillin and penicillin has been described previously.26 In addition, Chajçcka-Wierzchowska et al.8 reported that isolates with resistance to methicillin, oxacillin and penicillin were also resistant to rifampicin, clindamycin and tetracycline. In this study, 33.3% of the eight methicillin-resistant staphylococci were also resistant to rifampicin, 25% to tetracycline and clindamycin and 25% to erythromycin.
Two mechanisms of tetracycline resistance have been identified in Staphylococcus spp. and are mediated by tet genes: plasmid-mediated tetK and tetL encoding efflux and tetM encoding ribosomal protection mediated determinants.27 In the current study, two isolates harbouring tetL and tetK were found to be resistant to oxytetracycline; in addition, one isolate was found to be resistant to oxytetracycline without the presence of any of the tet genes tested for. Previous studies have found discrepancies between phenotypic expression and the presence of resistance genes.27 In comparison, AR to tetracycline was seen in 14.7% of staphylococci isolates from soft cheese19, 18.9% of staphylococci isolates from cow mastitis20 and 34.5% of isolates from food of animal origin, of which all isolates resistant harboured at least one tet gene8. Osman et al.14 found 68% AR to tetracycline from raw beef and chicken meat in Egypt, which was considerably higher than this and other studies. A conjugative transposon (Tn6079) is responsible for the spread of tetL27, whereas tetM can be transposon-located or chromosomal27. Therefore, the presence of tetL and tetM can indicate the potential for horizontal gene transfer between organisms.
Conclusion
In conclusion, a diverse number of Staphylococcus spp. were associated with mushrooms and mushroom handlers' hands. Antibiotic resistance of these mushroom- and hand-associated Staphylococcus spp. demonstrates a public health threat due to the potential of antibiotic gene transfer to medically important Staphylococcus spp. Moreover, opportunistic infection that might result due to an AR Staphylococcus spp. could lead to an infection that is difficult to treat. The presence of AR organisms adds to the general concern around the reservoir of AR on food products and food handlers' hands. Future research should determine the source of Staphylococcus spp. in production and on the product at the market end, in order to determine the specific risk to the final consumer. In addition, future research should also focus on ARG transfer and mechanisms in Staphylococcus species within the agricultural environment.
Acknowledgements
We thank Ms L. Pattyn for PCR analysis. The MALDI-TOF equipment was based on the research supported in part by the National Research Foundation (NRF) of South Africa (grant specific unique reference number (UID) 74426). The financial assistance of the NRF towards this research is hereby acknowledged. Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF. S.D. and W.R. acknowledge the Department of Science and Innovation-NRF Centre of Excellence in Food Security for financial assistance. G.V.R. acknowledges the University of Pretoria for financial assistance.
Competing interests
We have no competing interests to declare.
Authors' contributions
S.D.: Conceptualisation, research design and interpretation of data (conceptualisation), laboratory work, data collection and analysis (methodology and data collection), framework, final revision of manuscript, editing for intellectual content and submission (drafting or critically revising the manuscript), project management. W.R.: Research design (conceptualisation), field work, sampling, laboratory work, data collection and analysis (methodology and data collection), framework and writing of manuscript (drafting or critically revising the manuscript), project management. G.V.R.: Conceptualisation, research design and interpretation of data (conceptualisation), laboratory work, data collection and analysis (methodology and data collection), framework, final revision of manuscript, editing for intellectual content (critically revising the manuscript). E.d.P.: Conceptualisation, research design and interpretation of data (conceptualisation), data analysis (methodology and data collection), framework, final revision of manuscript, editing for intellectual content and submission (drafting or critically revising the manuscript). L.K.: Research design (conceptualisation), data analysis (methodology and data collection), framework, final revision of manuscript, editing for intellectual content and submission (drafting or critically revising the manuscript), student supervision, project leadership, funding acquisition.
References
1. Noble W, Valkenburg H, Wolters C. Carriage of Staphylococcus aureus in random samples of a normal population. Epidemiol Infect. 1967;65(4):567- 573. https://doi.org/10.1017/S002217240004609X [ Links ]
2. Benjelloun Touimi G, Bennani L, Berrada S, Moussa B, Bennani B. Prevalence and antibiotic resistance profiles of Staphylococcus sp. isolated from food, food contact surfaces and food handlers in a Moroccan hospital kitchen. Lett Appl Microbiol. 2020;70(4):241-251. https://doi.org/10.1111/lam.13278 [ Links ]
3. Lowy F. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520- 532. https://doi.org/10.1056/NEJM199808203390806 [ Links ]
4. Gillespie B, Headrick S, Boonyayatra S, Oliver S. Prevalence and persistence of coagulase-negative Staphylococcus species in three dairy research herds. Vet Microbiol. 2009;134(1-2):65-72. https://doi.org/10.1016/j.vetmic.2008.09.007 [ Links ]
5. Osman K, Alvarez-Ordóñez A, Ruiz L, Badr J, ElHofy F, Al-Maary KS, et al. Antimicrobial resistance and virulence characterization of Staphylococcus aureus and coagulase-negative staphylococci from imported beef meat. Ann Clin Microbiol Antimicrob. 2017;16(1):35. https://doi.org/10.1186/s12941-017-0210-4 [ Links ]
6. Chajecka-Wierzchowska W, Zadernowska A, Nalepa B, Sierpi'Nska M, taniewska-Trokenheim t. Retail ready-to-eat food as a potential vehicle for Staphylococcus spp. harboring antibiotic resistance genes. J Food Prot. 2014;77(6):993-998. https://doi.org/10.4315/0362-028X.JFP-13-466 [ Links ]
7. Podkowik M, Bystroñ J, Bania J. Prevalence of antibiotic resistance genes in staphylococci isolated from ready-to-eat meat products. Pol J Vet Sci. 2012;15(2):233-237. https://doi.org/10.2478/v10181-011-0139-z [ Links ]
8. Chajçcka-Wierzchowska W, Zadernowska A, Nalepa B, Sierpinska M, Laniewska-Trokenheim L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin - phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015;46:222-226. https://doi.org/10.1016/j.fm.2014.08.001 [ Links ]
9. Virdis S, Scarano C, Cossu F, Spanu V Spanu C, De Santis EPL. Antibiotic resistance in Staphylococcus aureus and coagulase negative staphylococci isolated from goats with subclinical mastitis. Vet Med Int. 2010;2010:1-6. https://doi.org/10.4061/2010/517060 [ Links ]
10. World Health Organization. Antimicrobial resistance: Global report on surveillance. Geneva: World Health Organization; 2014. Available from: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf [ Links ]
11. Spellberg B, Gilbert DN. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014;59:S71-S75. https://doi.org/10.1093/cid/ciu392 [ Links ]
12. Rossouw W, Korsten L. Cultivable microbiome of fresh white button mushrooms. Lett Appl Microbiol. 2017;64(2):164-170. https://doi.org/10.1111/lam.12698 [ Links ]
13. ISO 18593. Microbiology of food and animal feeding stuffs-Horizontal methods for sampling techniques from surfaces using contact plates and swabs. Iso 18593 [webpage on the Internet]. c2004 [cited 2021 Jul 19]. Available from: http://www.iso.org/iso/catalogue_detail.htm?csnumber=39849 [ Links ]
14. Osman KM, Amer AM, Badr JM, Saad ASA. Prevalence and antimicrobial resistance profile of Staphylococcus species in chicken and beef raw meat in egypt. Foodborne Pathog Dis. 2015;12(5):406-413. https://doi.org/10.1089/fpd.2014.1882 [ Links ]
15. Standing T-A, Du Plessis E, Duvenage S, Korsten L. Internalisation potential of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar Typhimurium and Staphylococcus aureus in lettuce seedlings and mature plants. J Water Health. 2013;11(2):210. https://doi.org/10.2166/wh.2013.164 [ Links ]
16. Bauer A, Kirby W, Sherris J, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-496. https://doi.org/10.1093/ajcp/45.4_ts.493 [ Links ]
17. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. Annapolis Junction, MD: CLSI; 2018. [ Links ]
18. Nunes RSC, Pires de Souza C, Pereira KS, Del Aguila EM, Flosi Paschoalin VM. Identification and molecular phylogeny of coagulase-negative staphylococci isolates from Minas Frescal cheese in southeastern Brazil: Superantigenic toxin production and antibiotic resistance. J Dairy Sci. 2016;99(4):2641- 2653. https://doi.org/10.3168/jds.2015-9693 [ Links ]
19. Fontes CO, Silva VL, De Paiva MRB, Garcia RA, Resende JA, Ferreira-Machado AB, et al. Prevalence, antimicrobial resistance, and virulence characteristics of mecA-encoding coagulase-negative staphylococci isolated from soft cheese in Brazil. J Food Sci. 2013;78(4):594-599. https://doi.org/10.1111/1750-3841.12088 [ Links ]
20. Klimiene I, Virgailis M, Pavilonis A, Siugzdiniene R, Mockeliunas R, Ruzauskas M. Phenotypical and genotypical antimicrobial resistance of coagulase-negative staphylococci isolated from cow mastitis. Pol J Vet Sci. 2016;19(3):639-646. https://doi.org/10.1515/pjvs-2016-0080 [ Links ]
21. Graham JP Evans SL, Price LB, Silbergeld EK. Fate of antimicrobial-resistant enterococci and staphylococci and resistance determinants in stored poultry litter. Environ Res. 2009;109(6):682-689. https://doi.org/10.1016/j.envres.2009.05.005 [ Links ]
22. Wu S, Huang J, Zhang F, Wu Q, Zhang J, Pang R, et al. Prevalence and characterization of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China. Front Microbiol. 2019;10:1-13. https://doi.org/10.3389/fmicb.2019.00304 [ Links ]
23. Graham JP Price LB, Evans SL, Graczyk TK, Silbergeld EK. Antibiotic resistant enterococci and staphylococci isolated from flies collected near confined poultry feeding operations. Sci Total Environ. 2009;407(8):2701-2710. https://doi.org/10.1016/j.scitotenv.2008.11.056 [ Links ]
24. Papadopoulos P Papadopoulos T, Angelidis AS, Kotzamanidis C, Zdragas A, Papa A, et al. Prevalence, antimicrobial susceptibility and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus isolated from dairy industries in north-central and north-eastern Greece. Int J Food Microbiol. 2019;291:35-41. https://doi.org/10.1016/j.ijfoodmicro.2018.11.007 [ Links ]
25. Bunnueang N, Kongpheng S, Singkhamanan K, Saengsuwan P, Rattanachuay P, Dangsriwan S, et al. Methicillin-resistant Staphylococcus aureus from ready-to-eat foods in a hospital canteen, southern Thailand: Virulence characterization and genetic relationship. Southeast Asian J Trop Med Public Health. 2015;46(1):86-96. [ Links ]
26. Vyletelova M, Vlkova H, Manga I. Occurrence and characteristics of methicillin resistant Staphylococcus aureus and methicillin resistant coagulase-negative staphylococci in raw milk manufacturing. Czech J Food Sci. 2011;29:S11-S16. https://doi.org/10.17221/4443-CJFS [ Links ]
27. Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000;45(6):763-770. https://doi.org/10.1093/jac/45.6.763 [ Links ]
28. Martineau F, Picard FJ, Grenier L, Roy PH, Ouellette M, Bergeron MG. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT Trial. J Antimicrob Chemother. 2000;46:527-534. https://doi.org/10.1093/jac/46.4.527 [ Links ]
29. Strommenger B, Bartels MD, Kurt K, Layer F, Rohde SM, Boye K, et al. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J Antimicrob Chemother. 2014;69(3):616-622. https://doi.org/10.1093/jac/dkt413 [ Links ]
30. Rizzotti L, Simeoni D, Cocconcelli P Gazzola S, Dellaglio F, Torriani S. Contribution of enterococci to the spread of antibiotic resistance in the production chain of swine meat commodities. J Food Prot. 2005;68(5):955-965. https://doi.org/10.4315/0362-028X-68.5.955 [ Links ]
31. Jensen LB, Frimodt-Moller N, Aarestrup FM. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett. 1999;170(1):151-158. https://doi.org/10.1111/j.1574-6968.1999.tb13368.x [ Links ]
32. Böckelmann U, Dörries H, Neus Ayuso-gabella M, De Marçay MS, Tandoi V Levantesi C, et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl Environ Microbiol. 2009;75(1):154-163. https://doi.org/10.1128/AEM.01649-08 [ Links ]
33. Brosius J, Palmer M, Kennedy P, Noller H. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75(10):4801-4805. https://doi.org/10.1073/pnas.75.10.4801 [ Links ]
 Correspondence:
Correspondence:
Lise Korsten
Email: lise.korsten@up.ac.za
Received: 20 July 2020
Revised: 09 July 2021
Accepted: 23 July 2021
Published: 29 Nov. 2021
Editors: Pascal Bessong, Sandiswa Mbewana
Funding: National Research Foundation (UID 74426), DSI-NRF Centre of Excellence in Food Security, University of Pretoria














