Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.116 n.7-8 Pretoria Jul./Aug. 2020
http://dx.doi.org/10.17159/sajs.2020/7461
RESEARCH ARTICLES
Sequence-related amplified polymorphism markers - a tool for litchi breeders in Africa
Elliosha Hajari; Dzunisani Nonyane; Regina Cronje
Agricultural Research Council - Tropical and Subtropical Crops, Nelspruit, South Africa
ABSTRACT
Litchi represents an economically important crop in South Africa - however, the local industry is based on only five cultivars. In order to expand the gene pool and to extend the harvest season, new cultivars have been imported. Currently, cultivars are identified based on morphological characteristics, but these are not always reliable. Molecular markers provide a tool to supplement morphological characterisation, particularly in cases in which confusion exists. The present study reports on the application of sequence-related amplified polymorphism (SRAP) markers in litchi for assessment of genetic relationships and molecular characterisation. The results provide evidence for separation of cultivars based on maturation period and fruit characteristics. The SRAP markers provide a tool for molecular characterisation that can be readily used by researchers with limited budgets, which is common in many developing countries.
Significance: •We report on the application of SRAP markers as a tool for litchi breeders in resource constrained countries. •The tested molecular markers allowed for genotyping (molecular characterisation) of litchi cultivars and selections. •The markers also revealed relationships between genetic and morphological (phenotypic) characteristics
Keywords: SRAP markers, Litchi chinensis, genetic diversity, molecular characterisation, plant breeding
Introduction
Litchi (Litchi chinensis Sonn.) belongs to the Sapindaceae family and is a commercially important fruit tree in tropical and subtropical regions. It is indigenous to southern China, northern Vietnam and the Malay peninsula but is also cultivated in other countries including India, Taiwan, Thailand, Madagascar and South Africa.1,2 There is a long history of litchi cultivation globally, and accordingly, a large number of selections have been developed over time. In China, more than 500 litchi accessions are stored at the National Litchi Germplasm Gene Bank at the Institute of Fruit Tree Research in the Guangdong Academy of Agricultural Sciences. Litchi was introduced into South Africa in 1876 and cultivation currently occurs in subtropical, frost-free regions in Limpopo, Mpumalanga and KwaZulu-Natal Provinces.3 During the 2017/2018 season, 5545 tons of litchi were produced in South Africa, of which 65% was exported, 24% sold to the local market and 11% processed into products.3 Hence, litchi cultivation makes an important contribution to the gross domestic product of the country and contributes towards job creation in the agricultural and processing sectors.
Despite the availability of a variety of litchi cultivars, the South African industry is based on just five: 'Mauritius' (89.8% of plantings) and 'McLeans Red' (6.4%) with 'Wai Chee', 'Fay Zee Siu' and 'Third Month Red' making up the rest (3.8%). As a consequence, the local litchi industry is characterised by a short production season with a limited range of cultivars that have a narrow genetic base.4,5 In recent years, there has been increasing interest in expanding the gene pool of cultivars available in South Africa in order to extend the harvesting season (beyond the current window) to exploit the export market and to increase the genetic diversity available for breeding.6 The Agricultural Research Council'sTropical and Subtropical Crops (ARC-TSC) in Nelspruit has an active breeding programme aimed at developing new cultivars for the litchi industry. In order to expand the gene pool, 30 cultivars from other litchi-producing countries have been imported and are currently being evaluated under South African conditions. These cultivars (as well as others present in the gene bank) have to be accurately identified and characterised in order to verify the identity of cultivars and to ensure that inadvertent mix-ups are avoided, which can have devastating effects on the livelihoods of growers and costly legal implications (in terms of Intellectual Property Rights).
Currently, identification of litchi is based on morphological traits (including vegetative, floral and fruit characteristics). The International Union for the Protection of New Varieties of Plants (UPOV) outlines a set of morphological descriptors for identification of litchi cultivars based on the distinctness, uniformity and stability of physical traits.7 However, there are limitations to the use of these descriptors; for example, they can be inaccurate during the juvenile phase and can be influenced by environmental and other factors, which leads to misidentifications.8 This situation is further complicated by confusion surrounding the naming of litchi cultivars which occurred as cultivar names were translated from Cantonese and Mandarin during distribution to other countries.9,10 Furthermore, it is not uncommon for the same cultivar to have different names in various Chinese dialects.9 This confusion has led to homonymies (cultivars having the same name but with different genetic profiles) and synonymies (cultivars having different names but with identical genetic profiles) which are prevalent in litchi cultivation programmes globally. Dissemination of cultivars to other countries has amplified this problem.9 This confusion in litchi nomenclature is exacerbated by misidentification of seedlings and the observation that the same cultivar grown in different climates may produce fruit that appear morphologically different from that expected.11
An example of the confusion surrounding litchi nomenclature is exemplified by the Chinese cultivar 'Sanyuehong' which is known by its English translation of 'Third Month Red' in South Africa (the fruit of this cultivar mature in the third month of the lunar calendar in China, hence its name). This same cultivar is referred to by a third name 'Sum Yee Hong' in Australia.5 Other cultivars have slightly different spellings in different countries, as is the case for 'Feizixiao'/'Fay Zee Siu' and 'Nuomici'/'No Mai Chee' while others are pronounced similarly but spelled differently such as 'Huaizhi' and 'Wai Chee'.5
Considering the above, there is a need for a supportive tool that can overcome some of the limitations imposed by morphological identification. Molecular markers can be used for this purpose and to unravel the genetic relationships between cultivars. The advantages of molecular characterisation are that markers are stable, thereby allowing plants to be sampled at any developmental stage and they are not influenced by environmental, pleiotropic or epistatic effects.12 A range of molecular markers have been applied to litchi, each with advantages and limitations.13 Applications relating to the use of markers for genetic diversity assessment and verification of cultivar identity have dominated the literature, which is not surprising considering the confusion in litchi nomenclature.14 These include studies on isozymes10,15, random amplified polymorphic DNA (RAPD)16-20, amplified fragment length polymorphism (AFLP)21,22, simple sequence repeat (SSR)23-25, expressed sequence tagged SSRs (EST-SSRs)26,27, inter simple sequence repeat (ISSR)9 and single nucleotide polymorphism (SNP)28 markers.
However, none of the markers mentioned above specifically targets coding regions of the genome.29 One specific class of markers, the sequence-related amplified polymorphism (SRAP) markers, are novel because they target open reading frames. Furthermore, the unique construction of the primers, combined with the optimised polymerase chain reaction (PCR) running conditions, ensures efficient reproducibility.29 The SRAP markers have been used for a range of purposes, for example, assessment of genetic relationships in apple and related species in which separation of genotypes was found to be based largely on geographic distribution.30 Similarly, SRAP markers were used to analyse the genetic diversity of pomegranate31 and passion fruit32 with the former study reporting a low degree of genetic variation amongst genotypes while the latter reported a high degree of genetic variability. Other researchers have used a different approach, by linking SRAP markers to specific traits such as fruit shape in cucumber33 and the colour around the stone of peach34.
The aim of the present study was to investigate the suitability of SRAP markers for molecular characterisation and investigation of the genetic relationships between litchi cultivars maintained at the ARC-TSC (cultivars maintained in the gene bank and newly imported ones). An additional advantage of using SRAP markers is that their use requires simple laboratory equipment (the most complicated being a PCR machine), which makes this technology suitable for resource constrained researchers on the African continent and in other countries.
Materials and methods
Source material
Leaf samples (mature, hardened leaves from the most recent flush that were still soft at the time of collection) of 52 litchi cultivars (including all imported cultivars as well as those available in the gene bank) were collected at the ARC in Nelspruit, South Africa (-25.4884, 30.4028). Table 1 provides a summary of all the cultivars tested as well as their country of origin. Leaf samples were brought to the laboratory and immediately processed for analysis.
DNA extraction
Genomic DNA was extracted from leaf material using the Macherey-Nagel Kit (Düren, Germany) as per the manufacturer's instructions. Cell lysis was performed using a Precellys homogeniser with zinc zirconium beads (Bertin Technologies, France).
Polymerase chain reaction
All SRAP primers were first screened to ensure that clear peaks and non-ambiguous scoring data were obtained. The SRAP primers were chosen according to literature29, i.e. 16 combinations of ME1 - EM4 (details of the primers tested are provided in Table 2). Genomic DNA was amplified in 15 µL reactions containing 25 ng DNA, TaKaRa EmeraldAmp Max HS PCR master mix (TaKaRa, Shiga, Japan), and 0.2 µM forward and 0.2 µM reverse primers using a G-Storm thermocycler. The PCR amplification conditions were as follows: hot start denaturation at 98 °C for 1 min, followed by five cycles of 1 min of additional denaturation at 94 °C, 1 min of annealing at 35 °C and 1 min of elongation at 72 °C. The initial amplification was followed by 35 cycles of denaturation at 94 °C for 30 s, the annealing temperature was increased to 50 °C for 30 s and elongation occurred for 1 min at 72 °C followed by a final elongation for 5 min at 72 °C. The PCR products were visualised via capillary electrophoresis (Qiaxcel Advanced, Qiagen, Hilden, Germany) using OM500 running conditions. All reactions were repeated to verify data.
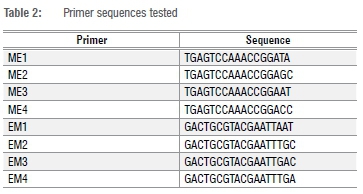
Data analysis
The sizes of the PCR products were determined using Screengel Software (Qiagen, Hilden, Germany) and the data were used to compile a database. A genetic distance matrix was created using GenAlEx 6.3, which was subjected to UPGMA (unweighted pair group method with arithmetic mean) cluster analysis. The genetic distance matrix was generated in a pairwise manner for each cultivar using the method of Huff et al.35 The information generated from the distance matrix provided a calculation of the differences between each pair of compared genotypes. The cluster analysis was validated through calculation of the cophenetic correlation coefficient (CCC) with branch lengths denoting genetic distances between samples. The final tree was constructed using MEGA (version 5.05).
Results and discussion
The present study reports on the use of SRAP markers to characterise litchi germplasm at the ARC-TSC, as well as newly imported cultivars that have not previously been investigated and promising selections arising from the local breeding programme. Examination of published literature indicates that there are only two reports on the use of SRAP markers in litchi.6,36 The first study considered the development of a core collection for breeding as the primary goal and the second study used different cultivars from the present investigation (cultivars of relevance to the Chinese industry). Furthermore, the current analysis included recently imported cultivars such as 'Baitangying', 'Jean Hang', 'Chompogo', 'Shujinqui', 'Maguili', 'Erdon Lee', 'Goose Egg', 'Yellow Red', 'Kwai May Red', 'Kim Cheng Meesa' and 'Hung Long', which have not been previously described in South Africa.
A summary of the band sizes attained with each of the primer pairs is provided in Table 3. All primer pairs were polymorphic to differing extents, as similarly reported by Bhatt et al.37 using SRAP markers in cumin. A total of 3736 bands were scored across all 16 primers. This figure is similar to the 3939 bands that were scored by Zhou et al.36 using 32 litchi genotypes and nine primer pairs. The total number of bands scored for each primer pair ranged from 77 with ME3/EM4 to 383 with ME2/EM1. Table 3 also highlights the most common band size present across all cultivars for each primer pair. This ranged from 252 bp with ME3/EM1 (present in 89% of the tested samples) to 1518 bp with ME3/EM2 (present in 60% of the tested samples).
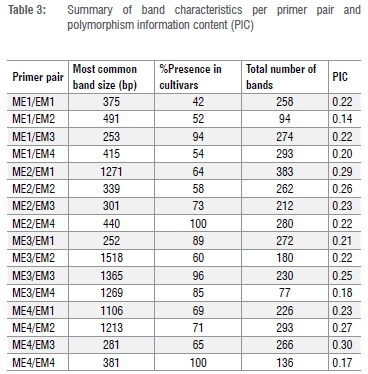
The polymorphism information content (PIC) was used to indicate the ability of polymorphic loci to reveal genetic diversity amongst genotypes and is also presented in Table 3.31 The PIC was lowest for ME1/EM2 (0.14) - this primer pair also generated fewer bands (94) than most of the other tested primers (>200). The highest PIC value of 0.30 was obtained with ME4/EM3. The average PIC in the present study (0.23) is similar to that reported by other researchers working on SRAP markers in various crops, e.g. 0.34 in cumin37, 0.28 in coffee38, 0.28 in pomegranate31 and 0.23 in passion fruit32. According to Xie et al.39, PIC values may be classified as revealing high (PIC>0.5), medium (0.5>PIC>0.25) or low (PIC<0.25) levels of polymorphism. In the present study, five primer pairs displayed a PIC value of >0.25 (indicative of medium levels of polymorphism between cultivars) while 11 had a PIC value of <0.25 (indicative of low levels of polymorphism between cultivars, Table 3). The low levels of polymorphism revealed by 11 of the tested primer pairs could be attributed to the high degree of genetic uniformity present among certain genotypes, for example, the 'Mauritius' selections and 'Madras' group (discussed below). Indeed, it is acknowledged that the commercially available litchi cultivars comprise a narrow genetic base23 as a result of selection for desirable fruit traits5. Furthermore, other commercially important tropical and subtropical crops (including citrus40 and mango41) are also characterised by having limited genetic variation.
The UPGMA dendrogram generated produced a CCC value of 0.945 with the Dice method indicating a good fit between the data and analysis method (Figure 1). The tested markers allowed for the separation of cultivars based on morphological characteristics and breeding history. For example, the five 'Mauritius' selections grouped together due to their high degree of genetic similarity. Similarly, 'Early Large Red' and 'Late Large Red' clustered together with a high degree of genetic similarity. 'Brewster' and 'Floridian' also clustered together and this is related to the fact that 'Floridian' is known to be the offspring of 'Brewster'. Further, these two cultivars display very similar morphological characteristics, which provides additional support for their close genetic relationship.9 These and other similar observations provided an indication that the SRAP markers were functioning effectively as the relationships observed could be explained in terms of known characteristics.
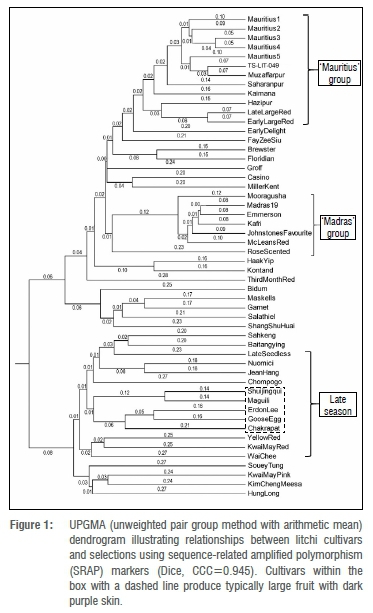
The cluster analysis revealed a number of groupings based on physical characteristics (highlighted in Figure 1). The first was the 'Mauritius' group comprising a cluster of genotypes that produce fruit resembling the cultivar 'Mauritius', i.e. large, ellipse-shaped fruit with a pointed to round tip, characterised by dull red skin and medium-sized seed. This is true for all cultivars within this group apart from 'Kaimana' which produces fruit that are different to 'Mauritius' (i.e. round fruit with flat sides, a round tip and bright red skin). Similarly, Degani et al.9 also reported the occurrence of a grouping of 'Mauritius'-like fruit using a different marker type, i.e. ISSR markers. In the present study, the occurrence of 'Kaimana' within the 'Mauritius' group might be explained by consideration of its parentage. 'Kaimana' is known to be an open pollinated seedling of 'Haak Yip' and it has been suggested that 'Mauritius' might be its other parent, thereby indicating a genetic relationship between 'Kaimana' and 'Mauritius'.42 'Early Delight' was also found to occur in close association with the 'Mauritius' group as the former was developed as the offspring from open pollinated 'Mauritius'6 and 'TS-LIT-049' is also a selection of 'Mauritius'.
As mentioned above, there is uncertainty surrounding the naming of litchi, which has been exacerbated by exporting cultivars to different countries and subsequent translations of cultivar names.9 Molecular markers can contribute towards dispelling some of this confusion. In this regard, 'Muzaffarpur' and 'Late Large Red' have previously been reported to be the same cultivar.43,44 However, Degani et al.9 demonstrated that these were different cultivars using ISSR markers. The results presented in the current study corroborate the findings of Degani et al.9 as differences were found between 'Muzaffarpur' and 'Late Large Red' (Figure 1). Furthermore, other cultivars often considered as synonyms (cultivars with different names but identical genetic profiles), e.g. 'Wai Chee'/'Salathiel' and 'Haak Yip'/'Souey Tung' were found to be different from each other in the present study, as evidenced from their positions on the dendrogram.23 Further evidence to support this assertion is provided by the observation that 'Salathiel' shared only 27.3% genetic similarity with 'Wai Chee' and 'Haak Yip and 'Souey Tung' shared 31.1% genetic similarity (as per the genetic distance matrix). A similar finding was reported by Viruel and Hormaza23 who used SSR markers. The above findings provide evidence that molecular markers can assist in efforts towards unravelling the confusion surrounding the naming of litchi cultivars. However, this needs to be a global effort involving all litchi-producing countries to ensure that standardised criteria are developed and applied consistently.
Examination of the cluster analysis revealed a second grouping of cultivars of Indian origin (also known as the 'Madras' group) which has been reported in other studies.9,25 Cultivars within this group were characterised by relatively short branch lengths (0.08-0.10; Figure 1) indicating close genetic similarity.45 These cultivars typically produce colourful, red fruit but under South African conditions, the fruit quality is often poor with soft, watery flesh and large seeds. In addition, alternate bearing can be a problem.46 However, these cultivars could prove to be useful breeding parents to transfer desirable attributes such as fruit colour to potential offspring.
Furthermore, a common problem in litchi production is cracking of the pericarp of fruit, which results in major losses to growers. Cracked fruit cannot be sold as fresh fruit and can only be used for juicing (provided that fungal contamination has not set in) which results in less income than does the sale of fruit. A number of factors contribute to fruit cracking, with cultivar-specific differences being one, thus indicating a genetic link. For example, 'Nuomici' is known to be prone to cracking, with up to 80% of fruit lost to cracking disorders.47 Considering this, cultivars within the 'Madras' group could also serve as a potential source of desirable traits for improved skin characteristics. In this context, the composition of the various cell layers comprising the pericarp is particularly important as this contributes significantly to the ability to resist the physical stresses associated with cracking.47 In addition, the ability to withstand cracking also promotes better post-harvest storage characteristics in terms of the ability of fruit to withstand storage diseases.47
Some of the groupings apparent on the dendrogram can be explained by consideration of morphological characteristics of the cultivars. For example, 'Bidum' and 'Haak Yip' occur in close association on the dendrogram (although in adjacent groups) and these two cultivars are reported to look very similar to each other apart from a few differences, i.e. fruit of 'Bidum' are slightly smaller with more variation in size, have yellow red markings on the skin rather than the purple red colour typical of 'Haak Yip', have more chicken tongue (shrivelled) seeds and are not as sweet as 'Haak Yip'.48 Chicken tongue seeds are a desirable trait as it means a larger percentage of flesh recovery than in fruit with larger seeds. 'Kwai May Red' and 'Kwai May Pink' also occur in close association on the dendrogram and the physical appearance of trees is also similar. It has been suggested that 'Kwai May Pink' might be a seedling of 'Kwai May Red'. Despite the high degree of genetic similarity (Figure 1) and similar tree morphological characteristics, there are slight differences in fruit characteristics of these two cultivars which allow for distinction between them. In this regard, 'Kwai May Red' has a red skin colour rather than the orange-pink colour of 'Kwai May Pink'. In addition, 'Kwai May Red' has firmer fruit with more small seeds, a higher flesh recovery and slightly better flavour than 'Kwai May Pink'. 5,48
The current analysis also revealed instances where reported parents and offspring occurred in different groups on the dendrogram. Such findings are not uncommon, as discussed below. For example, 'Yellow Red' is assumed to be the offspring of 'Brewster'; however, these occur in different groups on the dendrogram - a finding that was also reported by Degani et al.9 with ISSR markers. Hence, the parentage of 'Yellow Red' remains in question, as pointed out by those authors. Similarly, 'Sahkeng' is reported to be a seedling of 'Haak Yip' yet they occur in different groups. These two cultivars also display some differing morphological characteristics. For example, 'Sahkeng' produces short branches bearing large fruit with swollen skin segments and blunt protuberances, while 'Haak Yip' has long, fragile branches producing medium-sized fruit with smooth skin.5,48 Another example is 'Kaimana', which has been reported to be a seedling of 'Haak Yip' but they do not occur in close association in the present study. Similarly, Madhou et al.25 could not support 'Haak Yip' being the parent of 'Kaimana' when SSR markers were used. In the case of 'Salathiel', 'Nuomici' is reported to be its parent but they occur in adjacent groups on the dendrogram. These two cultivars also display fruit characteristics that are different from each other, for example, 'Salathiel' fruits are egg- to ball-shaped with thick, moderately rough skin while 'Nuomici' produces heart-shaped fruit with thin, smooth skin.5 Similarly, the results presented by Degani et al.15 using isozyme analysis could not support 'Nuomici' as the parent of 'Salathiel'.
Apart from a few exceptions, the general trend observed was that most of the cultivars that are harvested early and in the middle of the season occurred together at the top of the dendrogram while the late season cultivars occurred at the opposite end (Figure 1). Separation of cultivars based on maturation period has been reported using different markers, i.e. with RAPD and SNP markers.17,18,28 Considering this, it has been suggested that fruit maturation period should be considered as one of the primary factors in litchi taxonomy.9,23,25 A novel observation made in the present study is the presence of a sub-group within the late season cultivars which were characterised by production of large fruit with dark purple skin (Figure 1 - cultivars enclosed by the box with a dashed line). This is of interest locally, as these cultivars not only extend the harvest season, but also large fruit is a novel characteristic in the local market.
Conclusion
The current study reports on the suitability of SRAP markers for investigating the genetic relationships between litchi cultivars and selections. The tested markers allowed for separation of cultivars into groups based on similar fruit characteristics and fruit maturation period. Application of the SRAP markers allowed for creation of a molecular genotype reference database which will be a useful tool in the breeding programme in future. The imported cultivars expanded the gene pool available, particularly for the late season cultivars which were not previously available in South Africa (apart from 'Wai Chee'). Furthermore, many of the late season fruit are of a large size which is an additional benefit for exploitation in the local and international market. While other, more recently developed markers are available (e.g. SNP markers), they are significantly more expensive and require access to high-tech platforms for detection and analysis of data. Although the cost of DNA sequencing technologies is becoming more affordable in developed countries, this is not the case in developing countries where budgets for research are constrained. Hence, the SRAP markers provide an alternative tool to researchers lacking access to sophisticated platforms. The resource requirements are relatively simple and can be undertaken in a standard molecular biology laboratory.
Acknowledgements
The Agricultural Research Council (South Africa) and South African Litchi Growers' Association are acknowledged for funding and support.
Authors' contributions
All authors contributed to the conception and design of the study. Material preparation, data collection and sample analysis was performed by D.N. Data analysis, validation, data curation and writing was undertaken by E.H. R.C. contributed to data analysis and interpretation. All authors were involved in revisions of the manuscript and approved the final manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Data availability
The data for this study are not available because they include breeding germplasm that have Intellectual Property restrictions.
References
1.Menzel CM, Simpson DR. A description of lychee cultivars. Fruit Var J. 1990;45:45-56. [ Links ]
2.Oosthuizen JH. Recommended cultural practices and research priorities for litchis in South Africa. S Afr Litchi Growers' Assoc Yearb. 1993;5:2-5. [ Links ]
3.National Agricultural Marketing Council. South African Fruit Trade Flow. 2018;30:1-13. [ Links ]
4.Begemann GJ. The South African Litchi Industry - an overview. Acta Hort. 2014;1029:35-39. [ Links ]
5.Menzel CM, Huang X, Liu C. Cultivars and plant improvement. In: Menzel CM, Waite GK, editors. Litchi and longan: Botany, production and uses. Cambridge: CABI Publishing; 2005. p. 59-86. [ Links ]
6.Froneman IJ, Nonyane D, Severn-Ellis A, Cronje RB, Sippel A. Expanding genetic diversity of the South African litchi germplasm collection to promote plant improvement. Acta Hort. 2015;1127:365-372. https://doi.org/10.17660/ActaHortic.2016.1127.57 [ Links ]
7.International Union for the Protection of New Varieties of plants (UPOV). UPOV code: Litchi. Guidelines for conduction of tests for distinctness, uniformity and stability. Geneva: UPOV; 2012. [ Links ]
8.Arias RS, Borrone JW, Tondo CL, Kuhn DN, Irish BM, Schnell RJ. Genomics of tropical fruit tree crops. In: Schnell RJ, Priyadarshan PM, editors. Genomics of tree crops. New York: Springer; 2012. p. 209-239. [ Links ]
9.Degani C, Deng J, Beiles A, El-Batsri R, Goren M, Gazit S. Identifying lychee (Litchi chinensis Sonn.) cultivars and their genetic relationships using intersimple sequence repeat (ISSR) markers. J Am Soc Hort Sci. 2003;128:838-845. [ Links ]
10.Aradhya MK, Zee FT, Manshardt RM. Isozyme variation in lychee (Litchi chinensis Sonn.). Sci Hort. 1995;63:21-35. https://doi.org/10.1016/0304-4238(95)00788-U [ Links ]
11.Long Y, Cheng J, Mei Z, Zhao L, Wei C, Fu S, et al. Genetic analysis of litchi (Litchi chinensis Sonn.) in southern China by improved random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR). Mol Biol Rep. 2015;42:159-166. https://doi.org/10.1007/s11033-014-3755-8 [ Links ]
12.Agarwal M, Shrivastava N, Padh H. Review: Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008;27:617-631. https://doi.org/10.1007/s00299-008-0507-z [ Links ]
13.Pandey DK, Dey A, Singh J. Biotechnological advances in lychee (Litchi chinensis) and their future implication in improvement of crop. In: Kumar M, Kumar V, Prasad R, Varma A, editors. The lychee biotechnology. Singapore: Springer Nature; 2017. p. 59-100. https://doi.org/10.1007/978-981-10-3644-6 [ Links ]
14.Hajari E. Review: Molecular and related approaches to litchi improvement - historical perspective and future trends. J Hort Sci Biotech. 2019;94:693-702. https://doi.org/10.1080/14620316.2019.1624202 [ Links ]
15.Degani C, Beiles A, El-Batsri R, Goren M, Gazit S. Identifying lychee cultivars by isozyme analysis. J Am Soc Hort Sci. 1995;120:307-312. [ Links ]
16.Anuntalabhochai S, Chundet R, Chiangda J, Apavatjrut P. Genetic diversity within lychee (Litchi chinensis Sonn.) based on RAPD analysis. Acta Hort. 2002;575:253-259. https://doi.org/10.17660/ActaHortic.2002.575.27 [ Links ]
17.Liu C, Mei M. Construction of a lychee genetic linkage map based on RAPD markers. Acta Hort. 2003;625:131-136. https://doi.org/10.17660/ActaHortic.2003.625.13 [ Links ]
18.Liu C, Mei M. Classification of lychee cultivars with RAPD analysis. Acta Hort. 2005;665:149-160. https://doi.org/10.17660/ActaHortic.2005.665.17 [ Links ]
19.Kumar M, Gupta M, Shrivastava D, Prasad M, Prasad US, Sarin NB. Genetic relatedness among Indian litchi accessions (Litchi chinensis Sonn.) by RAPD markers. Int J Agric Res. 2006;1:390-400. https://doi.org/10.3923/ijar.2003.390.400 [ Links ]
20.Monshi FI, Bhuiyan MSU, Tabassum R. Adaptability of litchi germplasm in hilly areas of Sylhet Agricultural University and screening their genetic variation by using RAPD markers. Int J Plant Breed Genet. 2015;9:218-227. https://doi.org/10.3923/ijpbg.2015.218.227 [ Links ]
21.Yi G, Huo H, Chen D, Huang Z, Cai C, Qiu Y. Studies on genetic relationship among litchi varieties using AFLP. Acta Hort Sinica. 2003;30:399-403. [ Links ]
22.Pathak AK, Singh SP, Tuli R. Amplified fragment length polymorphism fingerprinting to identify genetic relatedness among lychee cultivars and markers associated with small-seeded cultivars. J Am Soc Hort Sci. 2014;139:657-668. https://doi.org/10.21273/JASHS.139.6.657 [ Links ]
23.Viruel MA, Hormaza JI. Development, characterisation and variability analysis of microsatellites in lychee (Litchi chinensis Sonn., Sapindaceae). Theor Appl Genet. 2004;108:896-902. https://doi.org/10.1007/s00122-003-1497-4 [ Links ]
24.Fan Q, Chen S, Zhou R, Shi S. Genetic variation of wild litchi (Litchi chinensis Sonn. subsp. chinensis) revealed by microsatellites. Conserv Genet. 2011;12:753-760. https://doi.org/10.1007/s10592-011-0182-4 [ Links ]
25.Madhou M, Normand F, Bahorun T, Hormaza JI. Fingerprinting and analysis of genetic diversity of litchi (Litchi chinensis Sonn.) accessions from different germplasm collections using microsatellite markers. Tree Genet Genome. 2013;9:387-396. https://doi.org/10.1051/fruits/2010009 [ Links ]
26.Xiang X, Chen DM, Ma SP, Ma WC, Fan J, Yang XY, et al. Core EST-SSR marker selection based on genetic linkage map construction and their application in genetic diversity analysis of litchi (Litchi chinensis Sonn.) germplasm resources. Acta Hort. 2014;1029:109-116. https://doi.org/10.17660/ActaHortic.2014.1029.12 [ Links ]
27.Sun Q, Shuaipeng MA, Liu W, Jiang N, Fang J, Yang X, et al. Identification and analysis on true or false hybrids from hybridisation populations of litchi (Litchi chinensis Sonn.) using EST-SSR markers. Agric Biotech. 2015;4:38-43. [ Links ]
28.Liu W, Xiao Z, Bao X, Yang X, Fang J, Xiang X. Identifying litchi (Litchi chinensis Sonn.) cultivars and their genetic relationships using single nucleotide polymorphism (SNP) markers. PLoS ONE. 2015;10, e0135390. https://doi.org/10.1371/journal.pone.0135390 [ Links ]
29.Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor Appl Genet. 2001;103:455-461. https://doi.org/10.1007/s001220100570 [ Links ]
30.Xu R, Hu D, Chen Z, Zhang C, Jiang X, Tang G. SRAP analysis on genetic relationships of genotypes in the genus Malus Mill. Biotech Biotech Equip. 2014;28:602-607. https://doi.org/10.1080/13102818.2014.948596 [ Links ]
31.Soleimani MH, Talebi M, Sayed-Tabatabaei BE. Use of SRAP markers to assess genetic diversity and population structure of wild, cultivated and ornamental pomegranates (Punica granatum L.) in different regions of Iran. Plant Syst Evol. 2012;298:1141-1149. https://doi.org/10.1007/s00606-012-0626-4 [ Links ]
32.Oluoch P, Nyaboga EN, Bargul JL. Analysis of genetic diversity of passion fruit (Passiflora edulis Sims) genotypes grown in Kenya by sequence-related amplified polymorphism (SRAP) markers. Ann Agrar Sci. 2018;16:367-375. https://doi.org/10.1016/j.aasci.2018.08.003 [ Links ]
33.Meng H, Chen S, Chen Z, Chai D, Li Y. SRAP markers for fruit shape in cucumber. Pakistan J Bot. 2012;44:1380-1384. [ Links ]
34.Han J-C, Liu G-J, Chang R-F, Zhang X-Z. The sequence-related amplified polymorphism (SRAP) markers linked to the colour around the stone (Cs) locus of peach fruit. Afr J Biotech. 2012;11:9911-9914. https://doi.org/10.5897/AJB12.396 [ Links ]
35.Huff, DR, Peakall R, Smouse PE. RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt) Engelm. Theor Appl Genet. 1993;86:927-934. https://doi.org/10.1007/BF00211043 [ Links ]
36.Zhou J, Fu J-X, Wu Z-X, Huang S-S, Zhang Y-F, Wang Y, et al. Genetic diversity in litchi and longan germplasm as determined by SRAP markers. Acta Hort. 2011;918:799-806. https://doi.org/10.17660/ActaHortic.2011.918.105 [ Links ]
37.Bhatt J, Kumar S, Patel S, Solanki R. Sequence-related amplified polymorphism markers based genetic diversity analysis of cumin genotypes. Ann Agrar Sci. 2017;15:434-438. https://doi.org/10.1016/j.aasci.2017.09.001 [ Links ]
38.Jingrade P, Huded AK, Kosaraju B, Mishra MK. Diversity genotyping of Indian coffee (Coffea arabica L.) germplasm accessions by using SRAP markers. J Crop Improv. 2019;33:327-345. https://doi.org/10.1080/15427528.2019.1592050 [ Links ]
39.Xie WG, Zhang XQ, Cai H, Liu W, Peng Y. Genetic diversity analysis and transferability of cereal EST-SSR markers to orchard grass (Dactylis glomerata L.). Biochem Syst Ecol. 2010;38:740-749. https://doi.org/10.1016/j.bse.2010.06.009 [ Links ]
40.Sagawa CHD, Cristofani-Yaly M, Novelli VM, Bastianel M, Machado MA. Assessing genetic diversity of Citrus by DArT_seqTM genotyping. Plant Biosyst. 2018;152:593-598. https://doi.org/10.3389/fpls.2017.00577 [ Links ]
41.Kuhn DN, Bally ISE, Dillon NL, Innes D, Groh AM, Rahaman J, et al. Genetic map of mango: A tool for mango breeding. Front Plant Sci. 2017;8:577. https://doi.org/10.1080/11263504.2017.1341438 [ Links ]
42.Husselman JH, Froneman IJ. Cultivars. In: De Villiers EA, Joubert PH, editors. The cultivation of litchi. Nelspruit: Agricultural Research Council - Institute for Tropical and Subtropical Crops; 2002. p. 31-49. [ Links ]
43.Singh LB, Singh UP. The litchi. Uttar Pradesh, India: Superintendent, Printing and Stationery; 1954. [ Links ]
44.Pandey RM, Sharma HC. The litchi. New Delhi: Indian Council for Agricultural Research; 1989. [ Links ]
45.McGarigal K, Cushman S, Stafford S. Cluster analysis. Multivariate statistics for wildlife and ecology research. New York: Springer; 2000. p. 81-127. [ Links ]
46.South African Department of Agriculture, Forestry and Fisheries (DAFF). Litchi production guideline. Pretoria: DAFF; 2013. Available from: https://www.daff.gov.za/Daffweb3/Portals/0/Brochures%20and%20Production%20guidelines/Production%20Guidelines%20Litchi.pdf [ Links ]
47.Huang X. Fruit disorders. In: Menzel CM, Waite GK, editors. Litchi and longan: Botany, production and uses. Cambridge: CABI Publishing; 2005. p. 141-151. [ Links ]
48.Singh G, Nath V, Pandey SR, Ray PK, Singh HS. The litchi. New Delhi: Food and Agriculture Organisation of the United Nations; 2012. [ Links ]
 Correspondence:
Correspondence:
Elliosha Hajari
Email: HajariE@arc.agric.za
Received: 08 Oct. 2019
Revised: 13 Mar. 2020
Accepted: 18 Mar. 2020
Published: 29 July 2020
Editor: Teresa Coutinho
Funding: Agricultural Research Council (South Africa), South African Litchi Growers' Association














