Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.115 n.11-12 Pretoria Nov./Dec. 2019
http://dx.doi.org/10.17159/sajs.2019/6362
RESEARCH ARTICLE
Identification of lactic acid bacteria and determination of selected biochemical properties in emasi and emahewu
Protus SimatendeI, II; Muthulisi SiwelaII; Tendekayi H. GadagaI
IDepartment of Environmental Health Science, University of Eswatini, Mbabane, Kingdom of Eswatini
IISchool of Agricultural, Earth and Environmental Sciences, University of KwaZulu-Natal, Pietermaritzburg, South Africa
ABSTRACT
Fermented foods are produced at household level for personal consumption in the Kingdom of Eswatini (formerly Swaziland). In this study, we determined the biochemical aspects, enumeration, isolation and identification of lactic acid bacteria (LAB) in emasi and emahewu - two Swazi traditional fermented foods. Emasi had an average pH of 4.68, titratable acidity of 0.9% and LAB count of 8.25 log CFU/mL. Emahewu had a pH of 3.62, titratable acidity of 0.4% and LAB count of 8.10 log CFU/mL. The LAB counts were consistent with observations for similar African fermented foods. The LAB from emasi and emahewu were identified through Gram stain, catalase reaction, sugar assimilation tests using API 50 CH test strips, and sequencing of 16S rDNA. It was found (from nine isolates) that Lactococcus lactis subsp. lactis and Leuconostoc mesenteroides were the common strains in emasi. Lactobacillus plantarum, Lactobacillus paracasei ssp. paracasei and Lactobacillus brevis were also detected. Lb. plantarum, L. mesenteroides ssp. mesenteroides, Lactobacillus fermentum, Lb. brevis, Wessella confusa, Lactobacillus acidophilus and Lb. lactis were found in emahewu (from 16 isolates). This finding was consistent with LAB found in a South African fermented milk, in which common genera were Leuconostoc, Lactococcus and Lactobacillus. Strains found in emahewu - mainly Lactobacillus spp., Weissella and Enterococcus - are similar to those found in ting, a South African fermented non-alcoholic beverage.
SIGNIFICANCE:
•This study provides the first documentation of microbial and biochemical aspects of the Swazi traditional fermented foods, emasi and emahewu.
Keywords: microbial, biochemical, Swazi traditional fermented foods, identity, fermenting bacteria
Introduction
Fermentation of food is one of the oldest forms of food preservation.1 Several studies have shown how this technique helps in preventing food-borne illnesses, including childhood diarrhoea.2 Consumption of fermented foods is thought to contribute to good health because of the benefit of their microflora to the human gut.3
Fermented foods can be grouped into four categories: alcohol, lactic acid, acetic acid and alkali fermented foods.1,4 Several traditional African fermented cereal grain foods, such as mahewu (sour sorghum or maize meal non-alcoholic beverage from South Africa, Zimbabwe and Lesotho), togwa (thin sour maize meal porridge from Tanzania), kenkey (thick sour maize meal porridge from Ghana), amasi (spontaneously fermented milk from southern Africa) and motoho (thin sour sorghum porridge or beverage from Lesotho), are largely products of lactic acid fermentation.
Lactic acid bacteria (LAB) have been found to be the predominant microorganisms in most of these products.1 However, yeasts are also important in alcoholic fermented foods4, and may be accidental contaminants in fermented milk5,6. Mathara et al.7 found that Lactobacillus species (Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus paracasei and Lactobacillus acidophilus) were predominant in kule naoto, Kenyan traditional fermented milk produced by the Maasai. Other genera isolated from kule naoto were Enterococcus, Lactococcus and Leuconostoc. Schoustra et al.8 reported that Lactobacillus and Weissella were common genera, together with Lactococcus, Streptococcus and Leuconostoc, in Munkoyo and Chibwantu, traditional non-alcoholic fermented beverages popularly consumed in Zambia. Emasi and emahewu are non-alcoholic lactic acid fermented traditional foods produced by households in Eswatini.
The preparation methods of sancta (fermented maize meal), incwancwa (fermented porridge), emasi (fermented milk), emahewu (non-alcoholic cereal beverage), umcombotsi (alcoholic sorghum or millet beverage), mankanjane (malt distilled spirit), buganu/marula wine and papaya beer (fermented fruit mashes) have been previously outlined.9 However, the microbial flora responsible for the fermentation has not been studied. The aim of this study, therefore, was to investigate the microbial diversity, to isolate potential probiotic LAB strains and to identify LAB in emasi and emahewu. Some of the biochemical properties were also investigated. This step is important in up-scaling and possible commercialisation of these products.
Materials and methods
Location of study
This study was done in the Hhohho Region of the Kingdom of Eswatini (formerly Swaziland). Hhohho Region is in the highveld of the country where the temperatures range from very cold to warm. The Region is divided into 14 local administrations called tinkhundla. Samples were collected from 5 tinkhundla that were randomly selected from the 14 tinkhundla using a lottery system.
Sample collection
Samples of emasi and emahewu were collected from nine locations within the five tinkhundla: Lobamba (coded L), Mangwaneni (M), Zone 4 (Z4), Motshane (Mot), Mbabane (Mb), Ezulwini (Ez), Mvutjini (Mv), Sitjeni (S) and Ntfonjeni (Nt) areas in Hhohho, Swaziland. At each inkhundla, a list of members of the community who were known to prepare fermented foods was compiled with the assistance of community leaders, such as the umphakatsi or schoolteachers. Samples of fermented food were then collected from the households that were randomly selected from the list. The samples were collected in sterile screw-capped bottles and ferried in a cooler box to the laboratory at the University of Swaziland (a distance of 5-75 km) for analysis.
Preparation of emahewu and emasi
Women who prepared emahewu explained that the product was prepared by thoroughly mixing 1 kg maize meal with 5 L of water. The mixture was then cooked until well gelatinised into a soft porridge called umhidvo, then cooled (to 25-30 °C) and left to ferment at room temperature (25-30 °C; 2-5 days). Malt was not added during preparation of the product, therefore emahewu lacked the enzymes that come with addition of malt to trigger the start of fermentation. However, some households reported adding sugar or a peeled potato, therefore some bacterial inoculum may have originated from addition of potato and/or from bacteria that may have been present on utensils used during preparation.
Emasi was prepared by leaving raw milk to naturally ferment at room temperature (25-30 °C; 2-3 days) using plastic or metal containers or clay pots. The whey was sometimes strained to give a thick product. Back-slopping - which is inoculation using substrate from a previous fermentation - is often used during fermentation of milk to emasi.
Determination of pH and titratable acidity
The pH was determined using a Hanna Instruments pH meter (HI 8314, Leighton Buzzard Bedfordshire, UK) after calibrating with buffers at pH 4 and pH 7. Titratable acidity (TA) was determined using standardised 0.1 N NaOH (Rochelle Chemicals, Johannesburg, South Africa) according to the Association of Official Analytical Chemists (AOAC) method no. 947.05.10
Microbiological analysis
Enumeration of LAB
The fermented samples were analysed immediately upon arrival at the laboratory. LAB were enumerated on De Man, Rogosa and Sharpe (MRS) agar (Oxoid, Basingstoke, UK; CM0361) (selective agar) by spreading 0.1 mL of appropriate serial dilutions and incubating anaerobically at 30 °C for 48 h. Anaerobic conditions were created using an Oxoid anaerobic gas generating system (Oxoid, Basingstoke, UK, BR0038B) according to the manufacturer's instructions.
Isolation and selection of LAB strains
Colonies with a different appearance (based on colour, shape and size) were extracted from the MRS agar and purified by streaking on a fresh MRS agar plate. The purification process was repeated until single colonies with distinct appearance were obtained. The pure isolates were tested for Gram and catalase reactions. Cell morphology was observed under the microscope. The isolates that were Gram positive and catalase negative were taken as presumptive LAB. The LAB isolates were stored at -20 °C in MRS broth (Biolab, Wadeville, South Africa; HG000C87.500) containing 20% (v/v) glycerol until required for further tests.
Identification of LAB using Analytical Profile Index kits
The frozen LAB isolates were thawed and resuscitated by inoculating into fresh MRS broth and incubating at 30 °C for 24 h. A portion of the fresh culture was streaked onto MRS agar, which was then incubated anaerobically for 48 h. The pure colonies were extracted and inoculated onto Analytical Profile Index (API) 50 CH (bioMerieux, Marcy l'Etloile, France; Ref 50 300) test strips according to the manufacturer's instructions. The sugar fermentation profiles were then used to identify the isolates using API identification software (APIWEBTM). A total of 16 LAB strains from emahewu and 9 LAB strains from emasi were identified using the API 50 CH kit. The carbohydrate profile was generated based on substrate metabolism using the API 50 CH kit. The API 50 CH approach is a well-established accurate method for manual microorganism identification for Gram-positive and Gram-negative bacteria and yeast to the species level based on extensive databases. The system offers a large and robust database accessible through the Internet-based APIWEBTM service. The method is economical to run and user-friendly.
Identification of LAB by sequencing 16S rDNA
Identification of LAB was performed at Inqaba Biotec Industries (Pretoria, South Africa). Briefly, DNA was extracted using ZR Fungal/Bacteria DNATM kit (Zymo Research, Irvine, CA, USA). The 16S rDNA target region was amplified using DreamTaqTM DNA polymerase (Thermo ScientificTM, Waltham, MA, USA) and the primers 16S-27F (sequence 5′-AGAGTTTGATCMTGGCTCAG-3′) and 16S-1492R (sequence 5′-CGGTTACCTTGTTACGACTT-3′). Polymerase chain reaction (PCR) products were gel extracted (Zymo Research, ZymocleanTM Gel DNA Recovery kit), and sequenced in the forward and reverse directions on the ABI PRISMTM 3500 x l Genetic Analyser. Purified sequencing products (Zymo Research, ZR-96 DNA Sequencing Clean-upTM kit) were analysed using CLC Main Workbench 7 followed by a BLAST search on the database of the US National Center for Biotechnology Information.11 Of the 16 LAB strains initially identified from emahewu using the API 50 CH kit, 9 were identified using the 16S rDNA method.
Statistical analysis
Mean (± standard deviation) was calculated for the pH, TA and microbial counts for the samples in the different categories using Microsoft Excel. The statistical significance (p<0.05) of the data sets was evaluated using Statistical Package for Social Science (SPSS) software.
Results and discussion
Emasi
pH and TA
The average pH of emasi was 4.68±٠.٢٥, and TA was 0.9±٠.08% (Table 1), which corresponds well with values obtained in other studies for naturally fermented milk. For example, Kebede et al.12 reported that sethemi, South African naturally fermented milk similar to emasi, had pH values of about 4.1-4.3. Beukes et al.13 also reported that the pH of indigenous fermented milks from South Africa and Namibia ranged from 4.0 to 5.4, with an average of 4.6. Amasi produced at household level in Zimbabwe was found to have a mean pH of 3.98 and 1.0% TA.14 Gran et al.15 found that the pH of naturally fermented amasi produced by smallholder producers in Zimbabwe was about 4.2 after 48 h fermentation. Nunu is a Ghanaian spontaneously fermented milk with the consistency of yoghurt and a pH of about 3.4 after 48 h of fermentation.16 However, the reported TA of 4.5% for nunu was uncharacteristically high compared with the values recorded for emasi, amasi and other similar products in southern Africa. In comparison, Moyane and Jideani17 found that the pH of commercially produced amasi in Venda, South Africa, ranged from 4.22 to 4.34, with an average TA of 0.8%, which is close to what was recorded for spontaneously fermented emasi.
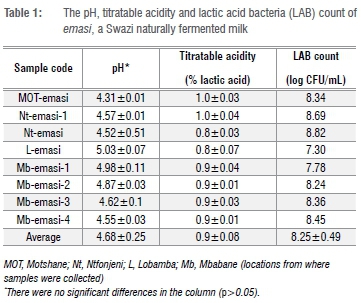
The pH range for emasi (Table 1) was 4.31-5.03. Although there were variations in pH of the samples, the deviations were not significant (p>0.05). The differences in varying values of pH in Table 1 may be attributed to the variations in the amount of available substrate for LAB to ferment, the type and quantity of predominant fermenting LAB (emasi production often involves back-slopping), and the duration of fermentation.
Enumeration of LAB
The LAB counts in emasi ranged from 7.30 to 8.82 log CFU/mL (translating to an average of 8.25±0.49 log CFU/mL) (Table 1). The LAB counts were very comparable to those of similar African naturally fermented milk products. For instance, the presumptive LAB counts in indigenous spontaneously fermented amasi from South Africa were about 7.7 x 108 CFU/mL (8.89 log CFU/mL).13 Zimbabwean amasi had a LAB ranging from 8.29 to 9.88 log CFU/g14, while a Nigerian fermented milk, nono, was found to have LAB counts of about 9.8 x 106 CFU/mL (6.99 log CFU/mL). In addition, Egyptian traditional fermented milk, Laban Zeer, had LAB counts of up to 7.4 log CFU/g. The Ghanaian nunu was also reported to have LAB counts of up to 9 log CFU/mL after 48 h fermentation.16 In contrast, Matsheka et al.18 reported a much lower value of 5.3 log CFU/mL LAB in madila, Botswanan spontaneously fermented milk.
Other studies on non-African fermented milks showed similar trends for LAB counts. Traditional naturally fermented goat's milk collected from households in the Haixi Region of China had LAB counts of 2.5 x 108-3.0 x 109 CFU/mL (8.4-9.5 log CFU/m).19
There was a relationship amongst the pH, TA and LAB of emasi. The LAB fermented the lactose in raw milk that led to production of organic acids. The organic acids lowered the pH and increased the TA. As the acidity in emasi increased over the processing time, it inhibited the growth of low tolerant LAB. The amount of fermentable lactose in raw milk therefore had an influence on pH and TA.
Emahewu
pH and TA
The average pH of emahewu was 3.61±٠.٥٥, ranging from 2.95 to 4.51. The TA was 0.42±0.17% (Table 2). A similar product prepared in Zimbabwe, which is also called mahewu, had a final pH of 3.0.20 This product had a TA of about 0.9% after 48 h fermentation, which is higher than that observed for emahewu. The Zimbabwean mahewu is made with maize meal and sorghum malt flour, which is probably the reason for production of higher amounts of organic acids. Sorghum malt is not added during preparation of emahewu.9 The pH in bushera, a non-alcoholic sorghum-based beverage from Uganda, was found to range from 3.7 to 4.5,21 which is close to the values obtained for emahewu. The TA of this product was 0.5%, which tallies with the results of the current study and the pH values obtained.
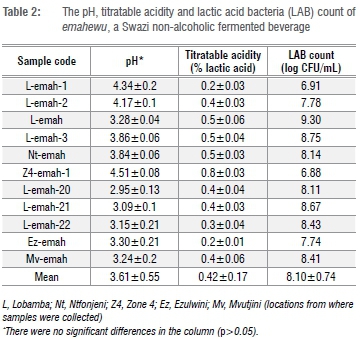
The pH range for emahewu (Table 2) was 3.09-4.51. Although there were variations in pH of the samples, the deviations were not significant (p>0.05). The differences in varying values of pH in Table 2 may be attributed to the variations in the amount of available substrate for LAB to ferment, the type and quantity of predominant fermenting LAB, and the duration of fermentation.
Enumeration of LAB
The LAB counts in emahewu ranged from 6.88 to 9.30 log CFU/mL (translating to an average of 8.10±0.74 log CFU/mL) (Table 2). The LAB counts were within the range expected when compared to those of other studies. Muyanja et al.21, in their study of bushera, found that the LAB counts varied between 7.1 and 9.4 log CFU/mL. LAB counts in homemade mahewu from Zimbabwe increased from 2.0 to 8.0 log CFU/mL after 72 h of fermentation.20Ting is a non-alcoholic beverage prepared in Botswana and is made from sorghum meal and malt. The LAB counts of ting were found to range between 8.08 and 10.1 log CFU/g.22
As with emasi, there was a relationship amongst the pH, TA and LAB of emhewu. The LAB fermented the carbohydrates (starch and some sugars) in maize meal used to make emahewu that led to production of organic acids. The organic acids lowered the pH and increased the TA. As the acidity in emahewu increased over the processing time, it inhibited the growth of low tolerant LAB. The amount of fermentable carbohydrates in maize meal therefore had an influence on pH and TA.
Identification of LAB
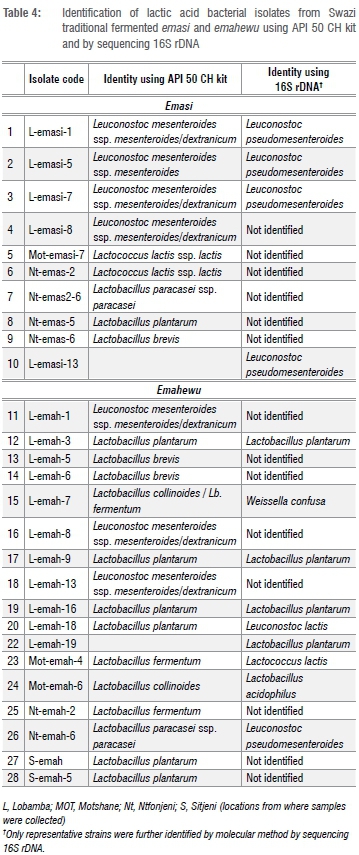
The isolates were initially screened as presumptive LAB using the Gram stain, catalase test and microscopic examination. The Gram-positive, catalase-negative isolates were identified to species level using API 50 CH test strips and by sequencing the 16S rDNA as shown in Table 3 and Table 4 and carbohydrate profile of LAB was as shown in Table 5.
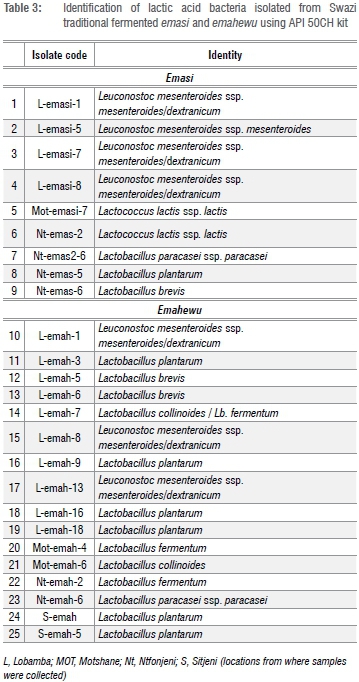
Emasi
Among the 9 emasi isolates identified using the API 50 CH kits, 4 were identified as Leuconostoc mesenteroides ssp. mesenteroides/dextranicum, 2 as Lactococcus lactis, 1 as Lb. plantarum and the other 2 as Lactobacillus brevis (Table 3). The four Leuconostoc isolates were characterised by sequencing the 16S rDNA and were identified as Leuconostoc pseudomesenteroides (Table 4), which was in close agreement with the API identification. The small difference in the identification of LAB between API and sequencing 16S rDNA methods is because the latter method is much more accurate than API.
In a study on South African naturally fermented milk, Beukes et al.13 reported that the genera Leuconostoc, Lactococcus and Lactobacillus were the main flora. The dominant lactococci species in the South African product was Lactococcus lactis subsp. lactis, while most of the Leuconostoc isolates were identified as Leuconostoc mesenteroides subsp. dextranicum, similarly to the findings for Swazi emasi. Other species identified in that study include Leuconostoc citreum, Leuconostoc lactis, Lactobacillus delbrueckii subsp. lactis and Lb. plantarum.
Mutukumira14 observed that Lactococcus lactis subsp. lactis was the predominant strain isolated from amasi, spontaneously fermented milk produced in Zimbabwe. The Zimbabwean amasi is produced in a similar way to emasi, which may explain the similarity in microbial ecology. Slight differences that may be found in the microbial diversity can be attributed to different types of containers used, as well as the environment under which the fermentation is done. Clay pots, metal containers, calabashes and gourds are often used and have been found to impact the microbial diversity.12 The current observations also agree with recent work by Osvik et al.23 who studied the bacterial diversity of amasi from the EkuPindiseni community of KwaZulu-Natal in South Africa using 16S rRNA and denaturing gradient gel electrophoresis for identification. The majority of the strains found were in the genus Lactococcus, as well as Lactobacillus, Leuconostoc and Enterococcus. However, a study by Mathara et al.7 showed that the genus Lactobacillus was predominant in kule naoto, Kenyan traditional fermented milk produced by the Maasai, in which the major Lactobacillus species was Lb. plantarum, followed by Lb. fermentum, Lb. paracasei and Lb. acidophilus. Other genera that were isolated in kule naoto were Enterococcus, Lactococcus and Leuconostoc.
Laban Zeer produced in Egypt seems to have similar flora to that of emasi. Saleh24 identified the LAB species in Laban Zeer as Leuconostoc mesenteroides subsp. cremoris, Lb. rhamnosus, Lb. plantarum, Lb. paracasei subsp. paracasei, Lb. delbruekii subsp. bulgaricus, Lb. curvatus subsp. curvatus and Lb. acidophilus. The most frequently isolated LAB species were found to be Leuconostoc mesenteroides subsp. cremoris and Lb. rhamnosus.
The Swazi fermented milk's microflora is therefore similar to that in other naturally fermented products from southern Africa, in particular amasi from South Africa and Zimbabwe, in which the dominant genera are Leuconostoc, Lactobacillus and Lactococcus.
Emahewu
Of the 16 isolates from emahewu identified using the API 50 CH test kit, 6 were identified as Lb. plantarum, 3 as Leuconostoc mesenteroides ssp. mesenteroides, 2 as Lb. fermentum, 2 as Lb. brevis, and 1 as Lb. collinoides (Table 3).
In comparison, the main LAB in ogi, a Nigerian fermented cereal beverage, were found to be Lb. plantarum, Lb. casei, Lb. brevis, Lb. fermentum, Lb. delbrueckii, Lb. acidophilus, Leuconostoc mesenteroides and Pediococcus acidilacti.25 In a separate study, Madoroba et al.26 isolated and identified LAB in ting, a South African spontaneously fermented sorghum non-alcoholic beverage, and found that the predominant LAB were Lb. plantarum, Lactococcus lactis, Lactobacillus fermentum, Lactobacillus rhamnosus, Weissella cibaria and Enterococcus faecalis. Some Enterobacteriaceae were also isolated. The Swazi emahewu samples were prepared from maize meal. The predominant microorganisms in koko, a Ghanaian spontaneously fermented porridge from millet, were identified as Weissella confusa and Lactobacillus fermentum27, while Yousif et al.28 found that Lactobacillus fermentum and Pediococcus acidilacti were the predominant strains in hussuwa, a Sudanese fermented sorghum food. Also, in gari, a cassava-based fermented food from Benin, Lb. plantarum was the most commonly isolated species followed by Leuconostoc fallax and Lactobacillus fermentum.29 Muyanja et al.21 also identified the LAB isolated from the spontaneously fermented Ugandan bushera as Lb. plantarum, L. paracasei subsp. paracasei, Lb. fermentum, Lb. brevis and Lb. delbrueckii subsp. delbrueckii. Similarly, Lactobacillus and Weissella were the common genera isolated from Munkoyo and Chibwantu, traditional non-alcoholic fermented beverages popularly consumed in Zambia.8 Therefore, the common LAB strains in Swazi emahewu belong to Lb. plantarum, Lactobacillus spp., Leuconostoc spp., Lactococcus spp. and Weissella spp. This finding is consistent with LAB strains reported in other products similar to Swazi emahewu. Notably, there were very few differences in identification of LAB for some isolates between the two methods (API 50 CH test and sequencing 16S rDNA). The accuracy of the API 50 CH test is limited to species available on the databases on the Internet-based APIWEBTM service, and the accuracy of 16S rDNA analyses strongly depends on the choice of primers.
Notably, the common LAB strains in Swazi emahewu belong to Lactobacillus, which suggests that Lb. plantarum, in particular, is a typical biota of spontaneously fermented maize and sorghum non-alcoholic beverages and plays a key role in defining the attributes of these products. Some strains of Lb. plantarum have been found to be amylolytic, that is, they break down starch in pearl millet slurries30; further studies on these emahewu strains is needed.
Carbohydrate profile of LAB
Almost all Lactobacillus spp. were able to utilise mainly ribose, galactose, D-glucose, D-fructose, D-mannose, N-acetylglucosamide, amygdaline, arbutine, esculine, salicine, cellobiose, maltose, lactose, melibiose, saccharose and trehalose (Table 5). Lactococcus ssp. metabolised carbon source ribose, D-xylose, galactose, D-glucose, D-fructose, D-mannose, N-acetylglucosamide, amygdaline, arbutine, esculine, salicine, cellobiose, maltose, lactose and trehalose. Most Leuconostoc mesenteroides ssp. utilised substrate ribose, galactose, D-glucose, D-fructose, D-mannose, cellobiose, maltose, lactose, melibiose, saccharose and trehalose. In general, LAB in the current study fermented other carbohydrates such as L-arabinose, rhamnose, mannitol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, melizitose, D-raffinose, starch, B gentiobiose, D-turanose, D-tagatose, D-arabitol, gluconate, 2-ketogluconate and 5-ketogluconate (Table 5).
The metabolism of carbohydrates by LAB is a similar observation to that made by Negussie et al.31 who observed that LAB isolated from Ethiopian naturally fermented buttermilk were able to utilise carbohydrates such as galactose, maltose, glucose, fructose, mannose and lactose. The results of the current study are supported by those of Ashmaig et al.32 who observed that LAB isolated from traditional Sudanese fermented camel's milk were able to ferment some carbohydrates. The common substrates that were fermented include carbohydrates such as lactose, fructose, galactose, trehalose, melibiose, mannose, xylitol and sorbose.
Conclusions
Emasi and emahewu are fermented foods of Swaziland. Leuconostoc mesenteroides, Lb. plantarum and Lb. lactis subsp. lactis were typical strains in emasi, while the Lactobacillus genus, especially Lb. plantarum, was typical in emahewu. Other LAB strains commonly found in emahewu were Lb. acidophilus, Leuconostoc lactis, Lactococcus lactis and Weissella confusa. Nevertheless, there is still a need to broaden the LAB isolates to be identified by sequencing 16S rDNA, carefully considering the choice of primers. Emasi and emahewu enhance dietary diversity and are popular foods for both children and adults in Eswatini. Studies are therefore needed to develop starter cultures for easier production of these foods.
Acknowledgements
We thank the University of Swaziland Research and the University of KwaZulu-Natal for funding part of the study, and Inqaba Biotec Industries (Pretoria, South Africa) for conducting the molecular sequencing to identify the lactic acid bacteria.
Authors' contributions
P.S. performed all the methodology, including data collection, sample analysis and data analysis as part of PhD studies; worked on the original concept of the manuscript write-up and revisions of the manuscript. M.S. and T.H.G. provided supervision and contributed significantly to the final version of the manuscript.
References
1.Blandino A, Al-Aseeri ME, Pandiella SS, Cantero D, Webb C. Cereal based fermented foods and beverages. Food Res Int. 2003;36:527-543. https://doi.org/10.1016/S0963-9969(03)00009-7 [ Links ]
2.Mortarjemi Y. Impact of small-scale fermentation technology on food safety in developing countries. Int J Food Microbiol. 2002;75:213-229. http://dx.doi.org/10.1016/S0168-1605(01)00709-7 [ Links ]
3.Gardiner G, Heinemann C, Baroja M, Bruce A, Beuerman D, Madrenas J, et al. Oral administration of the probiotic combination Lactobacillus rhamnosus GR-1 and L. fermentum for human intestinal applications. Int Dairy J. 2002;12:191-196. https://doi.org10.1016/S0958-6946(01)00138-8 [ Links ]
4.Steinkraus KH. Fermentation in world food processing. Compr Rev Food Sci Food Safety. 2002;1:23-32. http://dx.doi.org/10.1111/j.1541-4337.2002.tb00004.x [ Links ]
5.Gadaga TH, Mutukumira AN, Narvhus JA. The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int J Food Microbiol. 2001;68:21-32. https://doi.org/10.1016/S0168-1605(01)00466-4 [ Links ]
6.Roostita R, Fleet GH. Growth of yeasts in milk and associated changes to milk composition. Int J Food Microbiol. 1996;31:205-219. https://doi.org/10.1016/0168-1605(96)00999-3 [ Links ]
7.Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Holzapfel WH. Isolation, identification and characterization of the dominant microorganisms of kule naoto: The Maasai traditional fermented milk in Kenya. Int J Food Microbiol. 2004;94(3):269-278. https://doi.org/10.1007/s00284-007-9084-6 [ Links ]
8.Schoustra SE, Kasase C, Toarta C, Kassen R, Poulain AJ. Microbial community structure of three traditional Zambian fermented products: Mabisi, Chibwantu and Munkoyo. PLoS ONE. 2013;8(5), e63948, 12 pages. https://doi.org/10.1371/journal.pone.0063948 [ Links ]
9.Simatende P, Gadaga TH, Nkambule SJ, Siwela M. Methods of preparation of Swazi traditional fermented foods. J Ethnic Foods. 2015;2:119-125. https://doi.org/10.1016/j.jef.2015.08.008 [ Links ]
10.Association of Official Analytical Chemists (AOAC). Acidity of milk. 947.05 Method. Official methods of analysis of the Association of Official Analytical Chemists. 15th ed. Arlington, VA: AOAC; 1990. Available from: https://law.resource.org/pub/us/cfr/ibr/002/aoac.methods.1.1990.pdf [ Links ]
11.Altschul FS, Madden LT, Ffer SAA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-3402. https://doi.org/10.1093/nar/25.17.3389 [ Links ]
12.Kebede A, Viljoen BC, Gadaga TH, Narvhus JA, Lourens-Hattingh A. The effect of container type on the growth of yeast and lactic acid bacteria during production of Sethemi, South African spontaneously fermented milk. Food Res Int. 2007;40(1):33-38. https://doi.org/10.1016/j.foodres.2006.07.012 [ Links ]
13.Beukes EM, Bester BH, Mostert JF. The microbiology of South African traditional fermented milks. Int J Food Microbiol. 2001;63:189-197. https://doi.org/10.1016/S0168-1605(00)00417-7 [ Links ]
14.Mutukumira AN. Properties of amasi, a natural fermented milk produced by smallholder milk producers in Zimbabwe. Milchwissenschaft-Milk Science Int. 1995;50(4):201-205. Available from: https://eurekamag.com/research/009/259/009259407.php [ Links ]
15.Gran HM, Gadaga TH, Narvhus JA. Utilization of various starter cultures in the production of Amasi, a Zimbabwean naturally fermented raw milk product. Int J Food Microbiol. 2003;88(1):19-28. https://doi.org/10.1016/S0168-1605(03)00078-3 [ Links ]
16.Akabanda F, Owusu-Kwartengi J, Glover RLK, Tano-Debrah K. Microbiological characteristics of Ghanaian traditional fermented milk product, nunu. Nature Sci. 2010;8(9):178-187. Available from: http://www.sciencepub.net/nature/ns0809/23_3095_ns0809_178_187.pdf [ Links ]
17.Moyane JN, Jideani AIO. The physicochemical and sensory evaluation of commercial sour milk (amasi) products. Afr J Food Sci. 2013;7(4):56-62. https://doi.org/10.5897/AJFS12.089 [ Links ]
18.Matsheka MI, Magwamba CC, Mpuchane S, Gashe BA. Biogenic amine producing bacteria associated with three different commercially fermented beverages in Botswana. Afr J Microbiol Res. 2013;7(4):342-350. https://doi.org/10.5897/AJMR12.1645 [ Links ]
19.Zhang WY, Yun YY, Sun TS, Menghe B, Zhang HP. Isolation and identification of dominant microorganisms involved in naturally fermented goat milk in Haixi region of Qinghai, China. Ann Microbiol. 2008;58(2):213-217. https://doi.org/10.1007/BF03175319 [ Links ]
20.Simango C. Lactic acid fermentation of sour porridge and mahewu, a non-alcoholic fermented cereal beverage. J Appl Sci Southern Afri. 2002;8(2):89-98. https://doi.org/10.4314/jassa.v8i2.16926 [ Links ]
21.Muyanja CMBK, Narvhus JA, Treimo J, Langsrud T. Isolation, characterisation and identification of lactic acid bacteria from bushera: A Ugandan traditional fermented beverage. Int J Food Microbiol. 2003:80(3):201-210. https://doi.org/10.1016/S0168-1605(02)00148-4 [ Links ]
22.Sekwati-Monang B, Gänzle MG. Microbiological and chemical characterisation of ting, a sorghum-based sourdough product from Botswana. Int J Food Microbiol. 2011;150(2-3):115-121. https://doi.org/10.1016/j.ijfoodmicro.2011.07.021 [ Links ]
23.Osvik RD, Sperstad S, Breines E, Hareide E, Godfoid J, Zhou Z, et al. Bacterial diversity of amasi, a South African fermented milk product, determined by clone library and denaturing gradient gel electrophoresis analysis. Afr J Microbiol Res. 2013;7(32):4146-4158. https://doi.org/10.5897/AJMR12.2317 [ Links ]
24.Saleh FA. Isolation and identification of microorganisms and antibacterial activity of Laban Zeer, an Egyptian traditional fermented milk product. Scientific J Microbiol. 2013;2(2):31-42. Available from: https://www.academia.edu/2956449/Isolation_and_identification_of_microorganisms_and_antibacterial_activity_of_Laban_Zeer_an_Egyptian_traditional_fermented_milk_product [ Links ]
25.Dike KS, Sanni AI. Influence of starter culture of lactic acid bacteria on the shelf life of agidi, an indigenous fermented cereal product. Afr J Biotechnol. 2010;9(46):7922-7927. https://doi.org/10.5897/AJB09.1203 [ Links ]
26.Madoroba E, Steenkamp TE, Theron J, Scheirlinck I, Cloete TE, Huys G. Diversity and dynamics of bacterial populations during spontaneous sorghum fermentations used to produce ting, a South African food. Syst Appl Microbiol. 2011;34:227-234. https://doi.org/10.1016/j.syapm.2010.11.016 [ Links ]
27.Lei V, Jakobsen M. Microbiological characterization and probiotic potential of koko and koko sour water, African spontaneously fermented millet porridge and drink. J Appl Microbiol. 2004;96:384-397. https://doi.org/10.1046/j.1365-2672.2004.02162 [ Links ]
28.Yousif NMK, Huch M, Schuster T, Cho G, Dirar HA, Holzapfel WH, et al. Diversity of lactic acid bacteria from Hussuwa, a traditional African fermented sorghum food. Food Microbiol. 2010;27(6):757-768. https://doi.org/10.1016/j.fm.2010.03.012 [ Links ]
29.Kostinek M, Specht I, Edward VA, Schillinger U, Hertel C, Holzapfel WH, et al. Diversity and technological properties of predominant lactic acid bacteria from fermented cassava used for the preparation of Gari, a traditional African food. Syst Appl Microbiol. 2005;28(6):527-540. https://doi.org/10.1016/j.syapm.2005.03.001 [ Links ]
30.Songre-Outtara LT, Mouquet-Rivier C, Humblot C, Rochette I, Diawara B, Guyot JP. Ability of selected lactic acid bacteria to ferment a pearl millet-soyabean slurry to produce gruels for complementary foods for young children. J Food Sci. 2010;75(5):M261-M269. https://doi.org/10.1111/j.1750-3841.2010.01640.x [ Links ]
31.Negussie G, Fetien A, Fekadu B. Biochemical and molecular identification and characterisation of lactic acid bacteria and yeasts isolated from Ethiopian naturally fermented buttermilk. J Food Sci Technol. 2016;53(1):184-196. https://doi.org/10.1007/s13197-015-2049-z [ Links ]
32.Ashmaig A, Hasan A, El Gaali E. Identification of lactic acid bacteria isolated from traditional Sudanese fermented camel's milk. Afr J Microbiol Res. 2009;3(8):451-457. Available from: http://www.academicjournals.org/app/webroot/article/article1380279644_Ashmaig%20et%20al.pdf [ Links ]
 Correspondence:
Correspondence:
Protus Simatende
psimatende@yahoo.co.uk
Received: 13 May 2019
Revised: 13 Aug. 2019
Accepted: 02 Sep. 2019
Published: 27 Nov. 2019
Editor: Teresa Coutinho
Funding: University of Swaziland, University of KwaZulu-Natal














