Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.115 n.11-12 Pretoria Nov./Dec. 2019
http://dx.doi.org/10.17159/sajs.2019/5996
RESEARCH ARTICLE
Bacteria and yeast isolation and characterisation from a South African fermented beverage
Sanchia S. MoodleyI, II; Nomusa R. DlaminiI; Lucia SteenkampI; Elna M. BuysII
IBiosciences, Council for Scientific and Industrial Research, Pretoria, South Africa
IIDepartment of Consumer and Food Sciences, University of Pretoria, Pretoria, South Africa
ABSTRACT
Spontaneous fermentation of motoho, a southern African non-alcoholic sorghum beverage, results in products with inconsistent microbiological and sensory quality. We aimed to identify the microorganisms involved in the fermentation of motoho by using culture-dependent techniques as well as culture-independent polymerase chain reaction (PCR) screening and matrix-assisted laser desorption/ionisation time-of-flight analysis (MALDI-TOF). Lactobacillus, Candida, Rhodotorula and Geotrichum species were identified. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to evaluate the protein profiles of the isolated Lactobacillus species which produced protein bands of 14 kDa to 160 kDa, similar to those of other lactic acid bacteria isolated from various foods. A sensory panel evaluated and found significant differences (p<0.05) between the mouth feel, aroma and flavour of the traditional and modified motoho, with the latter being preferred. The microorganisms identified in this study could be used as starter cultures to optimise upscaled production of motoho.
SIGNIFICANCE:
•Traditionally fermented products have variable quality and the microorganisms isolated in this study could be used to decrease the variability in this fermented sorghum beverage
Keywords: non-alcoholic fermented sorghum beverage, MALDI-TOF, PCR, SDS-PAGE
Introduction
Motoho is a traditionally fermented non-alcoholic sorghum beverage produced by the Sotho people of southern Africa using spontaneous fermentation. Traditional fermentation is an unrestrained process and depends upon microorganisms present in the raw material or utensils, or added as a starter culture, and often results in variations in the microbiological safety as well as the quality of products.1 This variability can be overcome with the use of starter cultures2 and through understanding the fermentation process so that end products with consistent quality can be produced3. The lack of durability in terms of shelf life as well as variability in the quality of traditionally fermented products could inconvenience the consumer and result in a decline in the purchasing of the product.4 It was presumed that the phenotypic and genotypic characterisation of motoho would indicate that yeasts and lactic acid bacteria (LAB) would be the principal microorganisms responsible for fermentation, as is the case with the majority of African fermented foods.4-9 Previous studies have included the preparation method of motoho10 as well as the inhibitory effects of motoho fermentation on pathogenic microorganisms such as Salmonella typhi, Salmonella sp., Escherichia coli 0126 and Shigella boydii.11 However, to our knowledge, this study is the first to isolate and characterise the microorganisms involved in the fermentation of motoho.
The aim of this study was to assist a small to medium enterprise (SME) owner with the upscaling of motoho production from household to more industrialised scale and also to assist with improving the shelf life and quality of the product so that market accessibility could be promoted. To achieve this, it was necessary to identify the microorganisms involved in the fermentation, using both phenotypic and genotypic methods. The identified microorganisms could then be used as potential starter cultures to decrease the variability during the upscaling of motoho production. Upscaling would provide more job opportunities and thereby uplift the local community in Welkom, South Africa, from which this SME operates.
Materials and methods
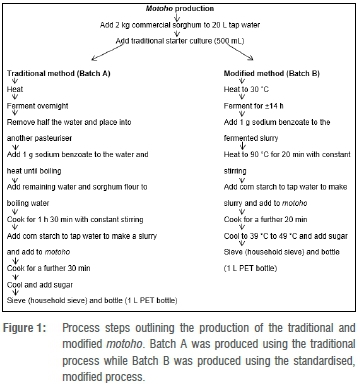
Traditional and modified production of motoho
Motoho was produced in the Department of Food Science pilot plant, University of Pretoria, South Africa, using traditional and modified methods and the methods described in detail in Figure 1. The modified motoho fermentation was prepared in triplicate (B, C and D) using the starter culture prepared from the back-slopping process, as described for the traditional process; and this preparation was added to a mixture of 2 kg sorghum flour in 20 L tap water. The mixture was added to a pasteuriser, heated to 30 °C and left to ferment for 14 h. This was followed by the addition of sodium benzoate and heating of the slurry to 90 °C with constant stirring for 20 min. The corn starch slurry was prepared in cold water and was added to the boiling mixture, followed by boiling for a further 20 min. The boiled product was cooled to temperatures of 39-49 °C, followed by the addition of sugar and mixing. The product was sieved using a hand-held household sieve and bottled in 1-L polyethylene terephthalate (PET) bottles. The bottled motoho products were stored at 4 °C and a shelf-life study was undertaken during 5 weeks of storage. Samples were taken at each process point during the manufacture of motoho for measurement of pH (Crison, Barcelona, Spain) and microbial evaluation.
Microbial enumeration, isolation and primary phenotypic characterisation
Triplicate samples of liquid suspension were collected aseptically at each process point for microbial analysis. The collection was done at the following stages: (1) sorghum + water, (2) sorghum + water heated to 30 °C, (3) fermented slurry, (4) heated and cooled sample, (5) sample after adding sugar and (6) bottling.
A mass of 5 g of each sample was diluted with 45 mL of sterile Maximum Recovery Diluent (MRD) consisting of 8.5 g NaCl (Merck, Darmstadt, Germany) and 1 g bacteriological peptone (Oxoid, Basingstoke, United Kingdom) per litre of distilled water. This solution was then serially diluted using MRD and the dilutions were surface inoculated onto selective agar plates as follows: Tryptone Soy Agar (Oxoid) for the total plate counts of all viable mesophilic aerobic organisms and de Man, Rogosa and Sharpe Agar (MRS; Merck) for the isolation of LAB. The plates were incubated at 30 °C for ٤٨ h. Yeasts were quantified on Potato Dextrose Agar (Merck) which was acidified using 10% tartaric acid. The plates were incubated for 120 h at 25 °C. The most probable number method was selected to test for the presence of E. coli using test tubes containing Lauryl Tryptose Broth (Oxoid). Tubes were incubated at 37 °C for ٢٤ h and thereafter checked for the presence of gas production.
A total of 24 colonies of both yeasts and LAB were randomly selected from the Potato Dextrose Agar and MRS plates, respectively, that displayed counts of between 30 and 300 colony forming units (CFUs) and sub-cultured to obtain pure cultures.
Preparation of pure cultures for characterisation
For characterisation through polymerase chain reaction (PCR), pure cultures of LAB were inoculated into 10 mL of MRS Broth (Merck) and incubated at 30 °C for 72 h with agitation at 80 rpm in a New Brunswick 25D incubator shaker (New Brunswick, NJ, USA). The yeast colonies were inoculated into 10 mL of Yeast and Mould Broth (5 g enzymatic digest of gelatine (Sigma Aldrich, St. Louis, MO, USA), 3 g malt extract (Merck), 12 g dextrose.H2O (BDH Chemicals, Poole, United Kingdom), 3 g yeast extract (Oxoid) in 1 L of purified H2O) and incubated at 25 °C for 120 h with agitation at 80 rpm (New Brunswick).
PCR analyses of LAB and yeasts isolated from motoho
Extraction of LAB and yeast DNA
The LAB pure cultures inoculated in MRS Broth and incubated for 72 h were centrifuged at 17 000 x g for 30 min to obtain cell pellets. The supernatant was discarded, the cell pellets were frozen in liquid nitrogen and then ground to a fine powder. DNA extraction was then performed according to the method of Sakallah et al.12 Yeast cell pellets were prepared from the pure yeast cultures that had been inoculated and incubated in Yeast and Mould Broth, as described, and the pellets were obtained by centrifugation at 17 000 x g for 30 min. The resulting pellets were washed once with 100 µL of a 0.1% sarkosyl (Sigma Aldrich) solution in distilled water and centrifuged again at 17 000 x g for 10 min and the sarkosyl discarded. The pellets were then frozen with liquid nitrogen and ground to a fine powder. Extraction of yeast DNA was performed according to the method of Rivas et al.13
PCR of isolated yeast and LAB from motoho
All PCR reaction mixtures (total volume 25 μL) contained 12.5 μL DreamTaq Green PCR Master Mix (2X) (Thermo Scientific, MA, USA) which comprised 0.4 mM deoxynucleotide triphosphates, Taq polymerase, buffer and 4 mM MgCl2. Also added was 2.5 μL (1 μM) of the forward primer, 2.5 μL (1 μM) of the reverse primer, 2 μL of DNA (about 10 ng) and 5.5 μL nuclease free water. All PCR reactions were performed using an Applied Biosystems 2720 Thermocycler (Life Technologies, CA, USA).
The bacterial (LAB) 16S rRNA gene was amplified using the primers 27F modified (5'-AGAGTTTGATCMTGGCTCAG-3) and 1492R (5'-TACGGYTACCTTGTTACGACTT-3') according to the method described by Zhang et al.14 The universal primer N21 (5'-GGATCCGAGGGTGGCGGTTCT-3') was used to amplify yeast genomic DNA according to the method of Naumova et al.15 The reference strains used (obtained from the culture collection of the Council for Scientific and Industrial Research, Pretoria, South Africa) were Lactobacillus casei ATCC 7469 (Lb. casei) as a positive control for LAB, while Candida krusei and Rhodotorula species were used as positive controls for the yeasts.
Electrophoresis of LAB and yeast PCR products
Electrophoresis was performed for LAB and yeast PCR products obtained from the amplification of LAB using the 27F modified primer sequence and that of yeast using the N21 primer. Amplicons (10 µL of PCR products) of the resulting LAB, yeast and reference strains were subjected to electrophoresis on a 1.2% agarose gel containing 0.5 µg/mL ethidium bromide. A GeneRuler 1-kb molecular ladder was used (Thermo Scientific). Electrophoresis was performed in 1 x TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.3) (Biorad, CA, USA) using a current of 80 V. Gels were visualised via a UV transilluminator and images were acquired through a Syngene GBox Gel system (Syngene, Cambridge, UK) mounted with a camera (Syngene).
Matrix-assisted laser desorption/ionisation time-of-flight analysis
Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) analysis was undertaken for LAB and yeasts obtained from various stages of motoho production using traditional and modified production methods. A total of 24 yeast and LAB isolates were appropriated from each stage of motoho production and sub-cultured onto acidified Potato Dextrose and MRS Agars, respectively, to obtain pure cultures. After 24 h, colonies were smeared onto the MALDI-TOF stainless steel target plate as a thin film (Bruker Daltonics, Bremen, Germany) and allowed to air dry. A volume of 1 µL of matrix solution formulated using α-cyano-4-hydrocinnamic-acid (Bruker Daltonics), 500 µL acetonitrile (50%; (Sigma Aldrich), 75 µL ultra-pure water (47.5%) and 25 µL trifluoroacetic acid (25%; (Sigma) was applied to the colony smear and allowed to air dry. Dried formulations were subjected to 40 laser pulses on six different positions of the sample for 15 s. The resulting spectra were compared to the reference spectra of microorganisms that were already present in the MALDI-TOF database. The MALDI-Biotyper (Bruker Daltonics) software was used to analyse the captured spectra. A standard bacterial sample of E. coli was included to validate and calibrate each run.
Protein analysis using sodium dodecyl sulfate polyacrylamide gel electrophoresis
LAB isolates confirmed via MALDI-TOF were further subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis in order to verify their identification. Only the LAB isolates which were identified to the genus level using MALDI-TOF analysis were chosen for SDS-PAGE protein profiling.
Single colonies were inoculated into 50 mL of MRS broth and incubated for 72 h at 30 °C with continuous agitation at 80 rpm using a temperature-controlled benchtop shaker (New Brunswick). The MRS broth was centrifuged at 17 000 x g for 30 min and the resultant pellets were frozen with liquid nitrogen and ground to a fine powder with a pestle and mortar. A volume of 500 μL of protein extraction buffer (750 mM Tris-HCl, pH 8.0; 15% sucrose; 0.25% protease inhibitor mix) (Sigma Aldrich) and 1% mercaptoethanol was added and the ensuing suspension was centrifuged at 17 000 x g for 20 min. The supernatant containing total soluble protein was removed and placed into 2-mL Eppendorf tubes and stored at -20 °C.
Casting and running of SDS-PAGE
The casting and running of SDS-PAGE protein gels was performed according to the Laemmli method.16 The resolving gel comprised 15% T, 3.3% Cbis and 40% SDS (w/v) while the stacking gel consisted of 6% T, 3.3% Cbis and 40% SDS (w/v). The electrophoresis buffer used was 1 x Tris/Glycine/SDS buffer (Biorad). About 20 μg of protein from each sample was loaded onto gels using 4 x sample buffer.16 Samples were heated at 100 °C for 4 min, placed on ice to cool, pulsed to collect at the bottom of the tube and then loaded onto SDS-PAGE gels.
A PageRuler Prestained Protein Ladder (Thermo Scientific) was included in each gel to use to estimate the protein band size. A current of 120 V was used to conduct electrophoresis. Following three washes of 10 min each with sterile distilled water, proteins were stained for about 3 h or overnight by immersing the gels in PageBlue Protein Staining Solution (Thermo Scientific) with gentle agitation at room temperature. Gels were then washed repeatedly with sterile distilled water until all excess dye was removed and the gels scanned.
Shelf life of bottled motoho
The shelf life of bottled motoho stored at 4 °C was investigated over a 5-week period by determining the total plate counts of all viable mesophilic aerobic organisms determined using Tryptone Soy Agar (Oxoid) incubated at 30 °C for 48 h.
Sensory evaluation of traditional and modified motoho
A consumer panel was recruited at the University of Pretoria (Pretoria, South Africa) to evaluate the sensory differences between motoho produced using the traditional versus modified processes. The motoho was initially assessed for microbial quality and declared to be safe for consumption. The panel, which consisted of 21 men and 30 women between the age of 19 and 37 years, was selected based on their familiarity with fermented cereal products similar to motoho. The appearance, aroma, mouth feel, and flavour or overall impression (like or dislike) of the motoho samples were assessed. A nine-point hedonic scale was used to rate the products.17
All procedures performed in studies involving human participants were in accordance with the ethical standards of the University of Pretoria, Division of Food Science. The ethical clearance number for this study was EC100811-045.
Statistical analysis
A one-way analysis of variance with a significance level of p<0.05 was used18 for the microbiological and sensory analyses. Principal component analysis was used to classify the results from the sensory analysis according to selected descriptors using the PRINCOMP procedure of the SAS statistical software.
Results
Change in pH during manufacture of traditional and modified motoho
The initial pH of the traditional fermentation mixture was 6.6±0.2, which dropped overnight to 4.2±0.6. The initial pH of the motoho mixture from the modified process was 6.2±0.1, which dropped to 3±0.3 overnight. The pH of motoho from the modified process was therefore lower than that of the traditionally processed motoho.
Microbiological counts of motoho produced using two production methods
The total plate counts, LAB and yeast counts increased at each process point - for both the traditional and modified processes - from the time the sorghum was mixed with water, after heating the mixture and after fermentation. The counts then decreased after cooking (Table 1). The microbial levels - namely total plate counts, LAB and yeast counts - at each corresponding process point were not significantly different (p>0.05) between the traditional and modified motoho (Table 1).
PCR analysis of LAB and yeasts isolated from motoho
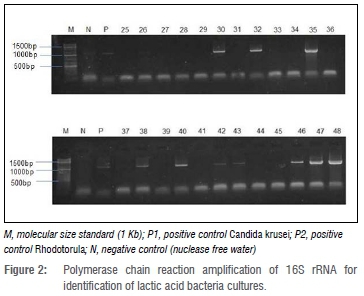
PCR amplification of 16S rRNA for identification of LAB cultures
The positive control Lb. casei showed similar banding patterns to 10 of the 24 selected isolates which also produced a band of between 1000 and 1500 base pairs (bp) and were thus considered most likely to be Lactobacillus strains. The agarose gel electrophoresis of the PCR amplification products with primers 27F and 1492R is illustrated in Figure 2.
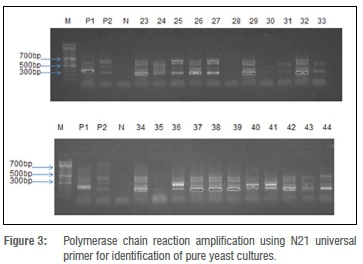
PCR amplification using the N21 primer for identification of yeast cultures
The primer N21 produced two characteristic bands of about 600 bp and 400 bp when assayed against the positive control C. krusei. The same primer produced four characteristic bands of 650 bp, 600 bp, 500 bp and 350 bp when Rhodotorula sp. was used as the positive control. Of the yeast isolates, 5 produced banding similar to C. krusei and 13 produced the four characteristic bands of Rhodotorula species. The agarose gel electrophoresis of the PCR products is illustrated in Figure 3.
MALDI-TOF identification of LAB and yeasts isolated from motoho
According to the standards recommended by the manufacturer, a log score of between 2.300 and 3.000 is considered a highly probable identification to species level while values of between 2.000 and 2.299 signify a secure classification to the genus level and a possible identification to the species level. Levels of 1.700 to 1.999 denote a likely classification to the genus level while values below 1.699 are regarded as not reliable.
In the current study, out of a total of 24 putative LAB isolates, 2 Lb. plantarum, 10 Lb. fermentum, 2 Lb. coryniformis and 4 Lb. paracasei were identified to the probable species level. Of the yeast isolates, 12 were identified as C. lambica, 4 were identified as C. kefyr, 1 as C. glabarata and 1 as C. pelliculosa; 1 R. mucilaginosa and 2 Geotrichum, namely G. candidum and G. silvicola, were also detected.
Protein characterisation of LAB using SDS-PAGE
Isolates identified as Lb. fermentum (Lanes 1, 3, 4, 5; Figure 4) showed similarities in protein band patterns. In Lane 1, two distinct bands (A and B) of between 10 and 25 kDa can be seen.
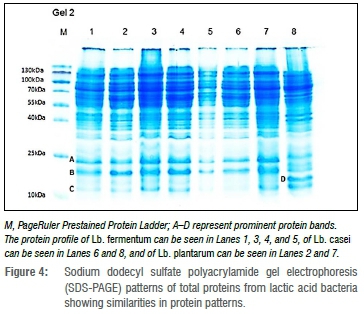
These bands are apparent throughout Lanes 1 to 7. A minor band of slightly more than 10 kDa is represented by C in Lane 1. This band can also be seen in Lanes 2 to 8. The protein profiles of Lb. plantarum are found in Lanes 2 and 7 while the profiles of the two Lb. paracasei isolates, seen in Lanes 6 and 8, differed slightly. A single prominent band of between 10 and 25 kDa is seen in Lane 8 (D) but not in Lane 7 nor in the other lanes.
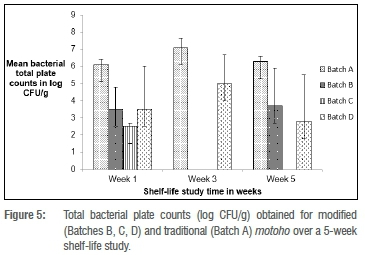
Shelf life of bottled motoho
The shelf life of motoho was assessed using total plate counts over a 5-week period; the results are shown in Figure 5. E. coli was not detected throughout the production process of motoho or during the 5-week shelf-life study.
Sensory analysis of traditional vs modified motoho
In the current study, there was no significant difference in the appearance of modified and traditional motoho (p>0.05). However, there were differences in aroma, mouth feel, and flavour or overall preference, with a preference for modified motoho (p<0.05). Figure 6 shows the principal component analysis of the sensory attributes of both the traditional and modified motoho.
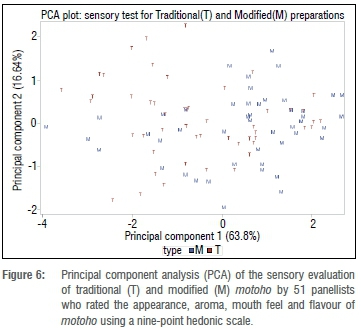
Principal component 1 (PC1) and principal component 2 (PC2) were extracted and accounted for 63.8% and 16.64% of the variance across the samples, respectively, within a four-variable system which included aroma, flavour, mouth feel and appearance. Along PC1, there is a separation between the traditional and modified motoho with the modified motoho appearing predominantly towards the right, indicating high levels for flavour and mouth feel, while the traditional motoho was found towards the left of PC1, showing a high intensity in the negative side for flavour and mouth feel. Along PC2 there appears to be an even distribution of both the modified and traditional towards the left and right sides, showing a high intensity in both the negative and positive sides in terms of appearance for both the traditional and modified motoho.
Discussion
This study is the first to evaluate the phenotypic, genotypic and sensory attributes of both traditional and modified motoho. Of the 24 LAB isolated from both the traditional and modified motoho, 10 were identified with 16S rRNA sequencing using the universal primers 27F and 1492R and produced bands of 1000-1500 bp, consistent with the band sizes generated by Zhang et al.14 from extracted bacterial DNA obtained from soil iron-manganese nodules. Nucleic acid purification can be challenging because foods, like motoho, are not simple matrices and the levels of DNA extracted from a mixed bacterial culture can also vary.19
Of the 24 yeasts that were isolated from motoho and amplified using the N21 universal primer, 75% showed positive amplification. Using the N21 primer, Naumova et al.15 also achieved effective amplification profiles of all 24 yeasts isolated from fermented sorghum beer.
No significant differences were found between the modified versus traditional production of motoho in terms of the yeast and LAB counts. However, simultaneous increases in the yeast and LAB counts were seen in the motoho produced by both methods. Mohamed et al.20 and Muyanja et al.21 obtained similar findings during the fermentation of kisra and togwa, respectively.
Yeasts and LAB are described as the principal microorganisms in fermented foods like ogi22-23, kisra20, hussuwa24 and togwa25. This is consistent with the microbial populations of both the traditional and modified motoho which showed the predominance of four LAB strains, namely Lb. fermentum, Lb. plantarum, Lb. coryniformis and Lb. paracasei, and yeasts C. lambica, C. glabarata, C. pelliculosa, C. kefyr, G. candidum, G. silvicola and R. mucilaginosa. The strains Lb. fermentum and Lb. plantarum were isolated from a number of products including obushera, a collection of traditionally fermented cereal beverages26, kenkey and ogi (fermented maize), fufu (fermented cassava) and kunun-zaki (fermented millet)27.Lb. paracasei forms part of the Lb. casei group and has been sequestered from fermented milk.28C. krusei, G. candidum and R. graminis were all isolated during the fermentation of maize for the production of ogi29, while R. mucilaginosa was isolated from bili bili, a traditional beer from Chad made from sorghum30.
Lactic acid bacteria that were isolated from motoho produced protein bands within the range of 14-160 kDa and showed similar protein banding patterns to LAB isolated from various food products, similarly to the study by Hébert et al.31 on regional cheese. Slight variations in protein profiles exist within each LAB species isolated from motoho. This variation could be ascribed to genomic heterogeneity of the different species, as was proposed by Sánchez et al.32 in relation to differences in the banding intensities of strains characterised as Lb. plantarum and Lb. paracasei, which were appropriated from sour dough33. The slight irregularities in the protein profiles could also be attributed to the differing origins, or process points of sampling the strains, as outlined by Pérez et al.34 with reference to a study conducted by Ghazi et al.35 Ghazi et al. aimed to identify the LAB, namely Leu. mesenteroides subsp. dextranicum, isolated from raw milk in Algeria using protein profiling, and also found differences in protein profiles.
Overall, the sensory profile (mouth feel, flavour and aroma) of the modified motoho was preferred over the traditional motoho. The higher yeast counts in the traditional motoho could have contributed to the lower sensory quality as yeasts are able to contribute to the organoleptic characteristics of the end products derived from fermentations.36
The microbiological counts of the motoho during the shelf-life study were unpredictable. This result could be because of the natural microflora of spontaneous food fermentations being uncontrollable, unpredictable and inefficient, or because they were destroyed by the heat treatments applied to the food.37 The modified motoho had lower microbiological counts over the 5-week period, possibly because the lower pH of the modified motoho may have had a preservation effect.38 Overall, the motoho microbial counts decreased for the modified process by Week 5, while there was a slight increase in microbial counts for the traditional motoho. These results are consistent with the results obtained in a shelf-life study on togwa25 in which the CFU/g decreased with increased storage time. A limitation of this study was that not all putative LAB isolates were identified using PCR and the low percentage of amplification of LAB DNA could be accounted for by the variability in DNA extraction.
Conclusion
The LAB and yeasts were identified to the species level using phenotypic and genotypic techniques. Lb. fermentum seemed to be the predominant microorganism during the production of motoho and will be characterised as a potential starter culture for future research on the large-scale production of motoho. Also, the modified method of motoho production resulted in a product with preferable sensory attributes and this method could be used during the upscaling of motoho. The modified motoho showed lower microbial counts than the traditional motoho throughout the 5-week shelf-life study and could have a longer shelf life. The results of this study may assist the SME in producing a more consistent product during upscaling. Future studies could include selecting specific microorganisms from those isolated and identified during this study to be used as starter cultures for the production of motoho.
Acknowledgements
We thank Mrs Rethabile Gladys Maimane from Golden Goose (Welkom, South Africa) for providing us with the traditional motoho recipe.
Authors' contributions
S.S.M. conducted the data analysis, collected the samples and wrote the initial draft. N.R.D. was responsible for the conceptualisation, assistance with methodology and writing revisions as well as student supervision and funding acquisition. L.S. assisted with writing revisions. E.M.B. was responsible for conceptualisation, assistance with methodology, student supervision, and writing revisions.
References
1.Nout MJ, Motarjemi Y. Assessment of fermentation as a household technology for improving food safety: A joint FAO/WHO workshop. Food Control. 1997;8(5-6):221-226. https://doi.org/10.1016/s0956-7135(97)00021-2 [ Links ]
2.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15(2):67-78. https://doi.org/10.1016/j.tifs.2003.09.004 [ Links ]
3.Marshall E, Mejia D. Traditional fermented food and beverages for improved livelihoods. Rome: Rural Infrastructure and Agro-Industries Division, Food and Agriculture Organization of the United Nations; 2011. Available from: http://www.fao.org/docrep/015/i2477e/i2477e00.pdf [ Links ]
4.Nout MJ. Ecology of accelerated natural lactic fermentation of sorghum-based infant food formulas. Int J Food Microbiol. 1991;12(2-3):217-224. https://doi.org/10.1016/0168-1605(91)90072-w [ Links ]
5.Halm M, Lillie A, Sørensen AK, Jakobsen M. Microbiological and aromatic characteristics of fermented maize doughs for kenkey production in Ghana. Int J Food Microbiol. 1993;19(2):135-43. https://doi.org/10.1016/0168-1605(93)90179-k [ Links ]
6.Hounhouigan DJ, Nout MJ, Nago CM, Houben JH, Rombouts FM. Changes in the physico-chemical properties of maize during natural fermentation of mawe. J Cereal Sci. 1993;17(3):291-300. https://doi.org/10.1006/jcrs.1993.1027 [ Links ]
7.Steinkraus KH. Classification of fermented foods: Worldwide review of household fermentation techniques. Food Control. 1997;8(5-6):311-317. https://doi.org/10.1016/s0956-7135(97)00050-9 [ Links ]
8.Nago MC, Hounhouigan JD, Akissoe N, Zanou E, Mestres C. Characterization of the Beninese traditional ogi, a fermented maize slurry: Physicochemical and microbiological aspects. Int J Food Sci Technol. 1998;33(3):307-315. https://doi.org/10.1046/j.1365-2621.1998.00169.x [ Links ]
9.Kunene NF, Geornaras I, Von Holy A, Hastings JW. Characterization and determination of origin of lactic acid bacteria from a sorghum-based fermented weaning food by analysis of soluble proteins and amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 2000;66(3):1084-1092. https://doi.org/10.1128/aem.66.3.1084-1092.2000 [ Links ]
10.Gadaga TH, Lehohla M, Ntuli V. Traditional fermented foods of Lesotho. J Microbiol Biotechnol Food Sci. 2013;2(6):2387. [ Links ]
11.Sakoane AL, Walsh A. Bacteriological properties of traditional sour porridges in Lesotho. In: Proceedings of a Workshop on Improving young child feeding in eastern and southern Africa: Household level food technology; 1987 October 11-16; Nairobi, Kenya. Ottawa: IDRC; 1988. [ Links ]
12.Sakallah SA, Lanning RW, Cooper DL. DNA fingerprinting of crude bacterial lysates using degenerate RAPD primers. Genome Res. 1995;4(5):265-268. https://doi.org/10.1101/gr.4.5.265 [ Links ]
13.Rivas R, Velázquez E, Valverde A, Mateos PF, Martínez‐Molina E. A two primers random amplified polymorphic DNA procedure to obtain polymerase chain reaction fingerprints of bacterial species. Electrophoresis. 2001;22(6):1086-1089. https://doi.org/10.1002/1522-2683()22:6<1086::aid-elps1086>3.0.co;2-6 [ Links ]
14.Zhang LM, Liu F, Tan WF, Feng XH, Zhu YG, He J. Microbial DNA extraction and analyses of soil iron-manganese nodules. Soil Biol Biochem. 2008;40(6):1364-1369. https://doi.org/10.1016/j.soilbio.2007.01.004 [ Links ]
15.Naumova ES, Korshunova IV, Jespersen L, Naumov GI. Molecular genetic identification of Saccharomyces sensu stricto strains from African sorghum beer. FEMS Yeast Res. 2003;3(2):177-184. https://doi.org/10.1016/s1567-1356(02)00191-5 [ Links ]
16.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680. https://doi.org/10.1038/227680a0 [ Links ]
17.Stone H, Sidel J. Sensory evaluation practices. Orlando, FL: Academic Press; 1985. [ Links ]
18.Fisher RA. XV: The correlation between relatives on the supposition of Mendelian inheritance. Earth Environ Sci Trans R Soc Edinb. 1919;52(2):399-433. https://doi.org/10.1017/s0080456800012163 [ Links ]
19.Ercolini D. PCR-DGGE fingerprinting: Novel strategies for detection of microbes in food. J Microbiol Methods. 2004;56(3):297-314. https://doi.org/10.1016/j.mimet.2003.11.006 [ Links ]
20.Mohammed SI, Steenson LR, Kirleis AW. Isolation and characterization of microorganisms associated with the traditional sorghum fermentation for production of Sudanese kisra. Appl Environ Microbiol. 1991;57(9):2529-2533. [ Links ]
21.Muyanja CM, Narvhus JA, Treimo J, Langsrud T. Isolation, characterisation and identification of lactic acid bacteria from bushera: A Ugandan traditional fermented beverage. Int J Food Microbiol. 2003;80(3):201-210. https://doi.org/10.1016/s0168-1605(02)00148-4 [ Links ]
22.Caplice E, Fitzgerald GF. Food fermentations: Role of microorganisms in food production and preservation. Int J Food Microbiol. 1999;50(1-2):131-149. https://doi.org/10.1016/s0168-1605(99)00082-3 [ Links ]
23.Blandino A, Al-Aseeri ME, Pandiella SS, Cantero D, Webb C. Cereal-based fermented foods and beverages. Food Res Int. 2003;36(6):527-543. https://doi.org/10.1016/s0963-9969(03)00009-7 [ Links ]
24.El Nour ME, El-Tigani S, Dirar HA. A microbiological study of Hussuwa: A traditional Sudanese fermented food from germinated sorghum bicolor cv feterita. World J Microbiol Biotechnol. 1999;15(3):305-308. https://doi.org/10.1023/a:1008849218617 [ Links ]
25.Mugula JK, Nnko SA, Narvhus JA, Sørhaug T. Microbiological and fermentation characteristics of togwa, a Tanzanian fermented food. Int J Food Microbiol. 2003;80(3):187-199. https://doi.org/10.1016/s0168-1605(02)00141-1 [ Links ]
26.Mukisa IM, Nsiimire DG, Byaruhanga YB, Muyanja CM, Langsrud T, Narvhus JA. Obushera: Descriptive sensory profiling and consumer acceptability. J Sens Stud. 2010;25:190-214. https://doi.org/10.1111/j.1745-459x.2009.00272.x [ Links ]
27.Steinkraus K. Handbook of indigenous fermented foods: Revised and expanded. New York: Marcel Dekker Inc.; 1995. [ Links ]
28.Kristo E, Biliaderis CG, Tzanetakis N. Modelling of rheological, microbiological and acidification properties of a fermented milk product containing a probiotic strain of Lactobacillus paracasei. Int Dairy J. 2003;13(7):517-528. https://doi.org/10.1016/s0958-6946(03)00074-8 [ Links ]
29.Omemu AM, Oyewole OB, Bankole MO. Significance of yeasts in the fermentation of maize for ogi production. Food Microbiol. 2007;24(6):571-576. https://doi.org/10.1016/j.fm.2007.01.006 [ Links ]
30.Maoura N, Mbaiguinam M, Nguyen HV, Gaillardin C, Pourquie J. Identification and typing of the yeast strains isolated from bili bili, a traditional sorghum beer of Chad. Afr J Biotechnol. 2005;4(7):646-656. https://doi.org/10.5897/ajb2005.000-3117 [ Links ]
31.Hébert EM, Raya RR, Tailliez P, de Giori GS. Characterization of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation. Int J Food Microbiol. 2000;59(1-2):19-27. https://doi.org/10.1016/s0168-1605(00)00282-8 [ Links ]
32.Sánchez I, Sese-a S, Palop L. Identification of lactic acid bacteria from spontaneous fermentation of 'Almagro' eggplants by SDS-PAGE whole cell protein fingerprinting. Int J Food Microbiol. 2003;82(2):181-189. https://doi.org/10.1016/s0168-1605(02)00260-x [ Links ]
33.Ricciardi A, Parente E, Piraino P, Paraggio M, Romano P. Phenotypic characterization of lactic acid bacteria from sourdoughs for Altamura bread produced in Apulia (Southern Italy). Int J Food Microbiol. 2005;98(1):63-72. https://doi.org/10.1016/j.ijfoodmicro.2004.05.007 [ Links ]
34.Pérez G, Cardell E, Zárate V. Protein fingerprinting as a complementary analysis to classical phenotyping for the identification of lactic acid bacteria from Tenerife cheese. Lait. 2000;80(6):589-600. https://doi.org/10.1051/lait:2000146 [ Links ]
35.Ghazi F, Henni DE, Benmechernene Z, Kihal M. Phenotypic and whole cell protein analysis by SDS-PAGE for identification of dominants lactic acid bacteria isolated from Algerian raw milk. World J Dairy Food Sci. 2009;4(1):78-87. https://doi.org/10.1016/j.fm.2003.11.006 [ Links ]
36.Romano P, Suzzi G, Domizio P, Fatichenti F. Secondary products formation as a tool for discriminating non-Saccharomyces wine strains. Antonie van Leeuwenhoek. 1997;71(3):239-242. https://doi.org/10.1023/a:1000102006018 [ Links ]
37.Giraffa G. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol Rev. 2004;28(2):251-260. https://doi.org/10.1016/j.femsre.2003.10.005 [ Links ]
38.Reis JA, Paula AT, Casarotti SN, Penna AL. Lactic acid bacteria antimicrobial compounds: Characteristics and applications. Food Eng Rev. 2012;4(2):124-140. https://doi.org/10.1007/s12393-012-9051-2 [ Links ]
 Correspondence:
Correspondence:
Sanchia Moodley
sanchia.moodley@gmail.com
Received: 30 Jan. 2019
Revised: 06 June 2019
Accepted: 25 July 2019
Published: 27 Nov. 2019
Editor: Pascal Bessong
Funding: European Union African Food Tradition Revisited by Research














