Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.115 no.9-10 Pretoria Set./Out. 2019
http://dx.doi.org/10.17159/sajs.2019/4771
RESEARCH ARTICLE
Detection of virulent thermophilic Campylobacter species in communal chickens
Marie E.A. BissongI, II; Collins N. AtebaI, III
IDepartment of Microbiology, School of Biological Sciences, North-West University, Mmabatho, South Africa
IIDepartment of Biomedical Science, University of Bamenda, Bambili, Cameroon
IIIFood Security and Safety Niche Area, Faculty of Agriculture, Science and Technology, North-West University, Mmabatho, South Africa
ABSTRACT
Campylobacter is a foodborne pathogen found in the gut of poultry and other animals, which thereby act as reservoirs for human infection. Campylobacter produces various virulence factors which contribute to the establishment of disease. The aim of this study was to detect virulence genes in Campylobacter isolated from chicken faeces. A total of 408 poultry cloacal samples were analysed through culture, PCR and DNA sequencing. Campylobacter were detected in 24 (5.9 %) of the samples, of which 21 were C. jejuni while 3 were C. coli. The cdtB gene was detected in all Campylobacter isolates while cdtA and cdtC genes were detected in 20 (83.3%) and 18 (75.0%) of the isolates, respectively. The cadF gene was detected in 18 (85.7%) C. jejuni but not in C. coli. Eight genotypes designated G1 to G8 were detected among the isolates. Genotypes G1 and G7 possessed all three cdt genes, a potential for producing functionally active toxins. A large proportion (70.8%) of the isolates possessed both genes for toxin production and adhesion. The high prevalence of cdt and cadF genes in Campylobacter species in this study indicates the pathogenic potential of the isolates. There is a need to implement control measures to curb the spread of these virulent strains.
SIGNIFICANCE:
•Virulent thermophilic campylobacters cause severe health complications in humans.
•Although chicken meat is the main source of human infections, there is limited data on the occurrence of Campylobacter in chicken faeces in southern Africa.
•This study adds to literature on the occurrence of virulent strains of Campylobacter species, notably through the deposition of the DNA sequences of our local strains into GenBank®.
•Communal chickens may serve as a source of virulent thermophilic campylobacters to humans, resulting in serious public health implications
Keywords: Campylobacter, virulence, genotype, prevalence, chicken
Introduction
Campylobacter species are Gram-negative curved bacteria which normally occur in the gastrointestinal tract of many domestic animals including poultry.1 These animals serve as potential reservoirs for human infection. If proper farm management techniques are not enforced, Campylobacter species can contaminate foods of animal origin and eventually gain access into the food chain.2-4 Undercooked meat, especially poultry, has been reported to serve as the main vehicle for the transmission of these organisms.5,6 Despite the fact that many species have been detected, it is the thermophilic species, most notably C. jejuni and C. coli, that are responsible for human infection.1 These species cause diseases in humans ranging from mild self-limiting diarrhoea to severe dysentery and, in some cases, the disease presents with severe complications such as Guillain-Barre Syndrome.7-9
Campylobacter virulence mechanisms include motility, mucus colonisation, toxin production, attachment, internalisation and translocation.10 These virulence factors and toxins are known to contribute to the survival and establishment of diseases in susceptible hosts.10 The flagella are used for motility of the bacterium and are known to enhance attachment of the bacterium to epithelial cells.7,11 The involvement of the bacterial flagella contributes to their pathogenicity in the host's gut.11 Invasiveness is also an important mechanism that enhances virulence in Campylobacter species.10 These bacteria become internalised into the sub-mucosal cells by means of the flagella and certain proteins.10 Invasion causes tissue damage and inflammation leading to gastroenteritis.10,12
Campylobacter adhesion to fibronectin (CadF) is an outer membrane protein which aids in the binding of C. jejuni and C. coli to intestinal epithelial cells,13 thus enhancing the colonisation of the gut. Bacterial toxins also play a key role in disease pathogenesis. Campylobacter species produce several cytotoxins, notably the cytolethal distending toxin (CDT) which causes direct damage to the DNA of the host cell and eventually leads to cell death.14 The CDT gene cluster consists of three protein subunits: cdtA, cdtB and cdtC. Although very little is known about the functions of the cdtA and cdtC gene fragments, cdtB has recently been identified to have nuclease activities.14 However, considering the fact that there are only a few Campylobacter genes that have been reported to be directly involved in virulence,15 the importance of the current study cannot be overemphasised.
Campylobacter is considered to be the most common cause of bacterial gastroenteritis worldwide.16 The rate of Campylobacter infections worldwide has been increasing and it is estimated that incidence of campylobacteriosis exceeds those of salmonellosis and shigellosis.17,18 This increase, as well as the expanding spectrum of diseases caused by Campylobacter species, necessitates a clearer understanding of the epidemiology and control of campylobacteriosis, especially in developing countries. Campylobacteriosis is common in Africa, especially among children under 5 years, with a prevalence of 7.7-21%.19-21
It is worth noting that the agropastoral industry is fast developing in South Africa in particular and in sub-Saharan Africa in general. Unfortunately, animals intended for human consumption are often natural reservoirs for pathogenic bacteria in humans. Infections with thermophilic Campylobacter present with severe complications in humans. Despite these health impacts, there are limited data on the prevalence of Campylobacter in clinical, environmental and food products in southern Africa.22-25 There is a need to exploit other sources, particularly animal species that are natural hosts of Campylobacter species, for the presence of potentially virulent strains. The present study was designed in order to isolate campylobacters from chicken cloacal samples and to determine the virulence profile of the isolates. Although other genes are responsible for the expression of pathogenicity in Campylobacter, the cdt gene subtypes (A, B and C) responsible for the expression of the cytolethal distending toxin and the cadF gene expressing products of adhesion were selected as targets for the present study.
Materials and methods
Study location
Samples were collected at two poultry farms in Mafikeng, North West Province of South Africa. Laboratory analyses were conducted in the Molecular Microbiology Laboratory of the Department of Microbiology, North-West University, Mafikeng Campus. Ethical clearance for the study was obtained from the Mafikeng Animal Research Ethics Committee (MAREC) of the North-West University, South Africa.
Sample collection
A total of 408 chicken cloacal swabs were collected using the culture swab transport system (COPAN, Italy) and transported on ice to the laboratory for analysis. The samples were analysed for the presence of Campylobacter species within 24 h.
Isolation of Campylobacter species
Each swab was rinsed in 5 mL of 2% (w/v) sterile peptone water (Biolab, Modderfontein, South Africa) and 100-µL aliquots spread-plated on Campylobacter selective agar supplemented with 5% (w/v) sheep blood and Campylobacter selective supplement (Merck, Darmstadt, Germany). The plates were incubated under microaerophilic (10% CO2) conditions at 42 °C for 24-72 h. Presumptive isolates were sub-cultured and stored in brain heart infusion broth with 20% (v/v) glycerol at -80 °C for further analysis.
Biochemical identification of Campylobacter species
All isolates were identified using standard biochemical tests.26 Isolates were Gram stained and observed for Gram-negative spiral rods. The oxidase test was performed using the oxidase test reagent obtained from Pro-Lab Diagnostics (Merseyside, UK) as indicated in the manufacturer's protocol. The catalase test was performed as previously reported.26 The Campylobacter test kit (Oxoid, Basingstoke, Hampshire, UK) was used to identify thermophilic campylobacters based on the manufacturer's description.
Molecular characterisation of Campylobacter species
Extraction of DNA
Pure isolates were inoculated into 10 mL of nutrient broth (Merck, Darmstadt, Germany) and incubated aerobically at 37 °C for ٢٤ h in a shaking incubator to enhance bacterial growth. Genomic DNA was extracted using the Zymo Research Genomic DNATM Tissue MiniPrep Kit (Zymo Research Corp, Irvine, CA, USA) based on the manufacturer's instructions and the samples were stored at -20 °C.
Detection of C. jejuni and C. coli
All isolates were screened for the presence of bacterial 16S rRNA gene fragments and identification of C. jejuni and C. coli was achieved using the hipO and asp gene primers, respectively (Table 1). Polymerase chain reaction (PCR) amplifications were performed using the model Bio-Rad C1000 TouchTM Thermal Cycler (Bio-Rad, Johannesburg, South Africa). The reactions were prepared in 25-μL volumes made up of 12.5 μL of One TaqR Quickload 2X Master Mix with standard buffer (Inqaba Biotec Ltd, Pretoria, South Africa), 1 μL of template DNA, 11 μL nuclease free sterile water and a final primer concentration of 0.2 μM. All PCR reagents were obtained from Inqaba Biotec Ltd (Pretoria, South Africa). PCR conditions comprised an initial denaturation at 95 °C for s min, followed by 35 cycles of denaturation at 94 °C for s, annealing at 55 °C for s and extension at 72°C for s. A final extension step was performed at 72 °C for 3 min. PCR products were stored at 4 °C until electrophoresis.
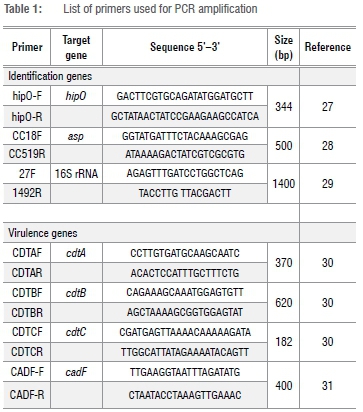
Detection of virulence genes
Virulence gene determinants associated with pathogenesis in Campylobacter species were determined by specific PCR analysis using the primers listed in Table 1. Cycling was performed using a DNA thermal cycler. The conditions for amplification were the same as those for the identification genes, except for cdtC and cadF genes, which were annealed at 50 °C.
Agarose gel electrophoresis
PCR products were resolved by electrophoresis on a 1% (w/v) agarose gel at 80 V for 3 h, using 1X TAE buffer (40 mM Tris, 1 mM EDTA and 20 mM glacial acetic acid, pH 8.0). A 100-bp DNA molecular weight marker (Fermentas, Glen Burnie, MD, USA) was included in each gel. The gels were stained in 0.001 µg/mL of ethidium bromide for 15 min and the amplicons visualised using a ChemiDoc Imaging System (Bio-RAD, Hercules, CA, USA).
DNA Sequencing
16S rRNA PCR amplicons were cleaned using a GeneJet PCR Purification kit (Thermo Fisher Scientific, Vilnius, Lithuania) and sequenced at Inqaba Biotec (Pretoria, South Africa). The identities of the isolates were confirmed through a BLAST search of the US National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results and discussion
Bacterial virulence is an important issue that needs scientific investigation. Several virulent determinants are associated with the survival and pathogenesis of Campylobacter species. This study was designed to determine the prevalence of Campylobacter species in chicken faeces and to identify virulence genes in the isolates. The cdt gene subtypes (A, B and C) responsible for the expression of the cytolethal distending toxin and the cadF gene expressing products of adhesion were selected for the study.
The occurrence of Campylobacter in both clinical and environmental samples from southern Africa has been reported.22-25 Diergaardt and colleagues reported the presence of Campylobacter spp. in both drinking and environmental water sources in Pretoria.22 In a study conducted in the Limpopo Province, Campylobacter species were found to be common causes of gastroenteritis in primary school children.23 Meanwhile, Uaboi-Egbenni and colleagues detected the presence of Campylobacter in goat faeces.32 In the Mafikeng area, Campylobacter has been detected in commercial milk as well as chicken meat.24,25 Despite these reports, information on the occurrence of virulent Campylobacter in chicken faeces in sub-Saharan Africa is scarce. In the present study, samples were collected from two commercial farms in Mafikeng. These farms sell live chickens to the local population and the chickens are slaughtered at the customers' homes. The handling of live chickens might enhance the spread of enteric pathogens including Campylobacter from chickens to humans, especially as standard hygiene practices might not be observed. In such scenarios, the presence of virulent campylobacters in chicken might present a serious public health hazard, more especially as these thermophilic bacteria can survive higher temperatures.
Isolation and molecular identification of Campylobacter species
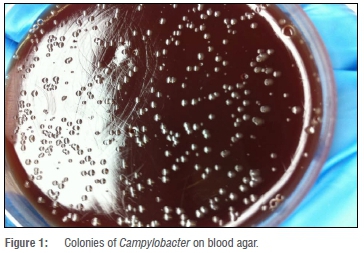
In this study, Campylobacter was isolated from 24 of the 408 (5.9%) chicken cloacal samples. However, of the samples inoculated, growth was observed in only 198 (48.5%), and out of this number, 126 (63.6%) were presumptively identified as Campylobacter based on growth on selective media and colony morphology. Isolates that presented with the characteristic mucoid cream to pink colonies (Figure 1) were tested using species-specific primers. The isolated DNA was tested for the presence of bacterial 16S rRNA gene fragments and all 126 (100%) presumptive isolates were positive - an indication that the genetic material was all from bacterial sources. Results of the PCR using specific primers showed that only 24 of 126 (19.0%) presumptive isolates were Campylobacter species; with 21 (16.7%) and 3 (2.4%) possessing the hipO and asp genes, respectively, specific for Campylobacter jejuni and Campylobacter coli, respectively (Figure 2).
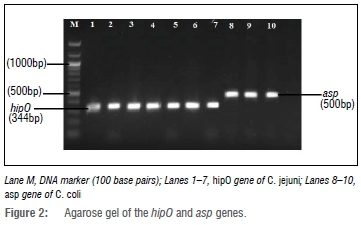
DNA Sequencing
Four isolates that were positive for the hipO gene were randomly selected and sent for DNA sequencing to confirm their identities. The identities of the isolates were confirmed through a BLAST search of the NCBI database and all the isolates were confirmed to be Campylobacter jejuni. In addition, one of the isolates that grew on the selective media but did not amplify with the campylobacter-specific primers was also sent for DNA sequencing using the universal 16S rRNA primer. This isolate was found to be Lactobacillus reuteri. Similar observations have been recorded in which isolates supposedly identified as Campylobacter, were later confirmed by PCR and or DNA sequencing to belong to other genera. In a related study, Diergaardt et al.22 confirmed by sequencing that only 3 out of 22 presumptive isolates were actually Campylobacter. In another study, out of 75 samples examined, only 1 sample (1.33%) was positive by cultural method while 5 samples (6.66%) showed the presence of Campylobacter species by PCR.33 Consequently, these reports confirm the superior efficacy of PCR over culture-based techniques for the rapid screening of both clinical and environmental samples. The sequences of the isolates were deposited in GenBank®, the sequence database of the NCBI, with the following accession numbers: MF872608 (Campylobacter jejuni, strain MOLcc14, 99% identity), MF872609 (Campylobacter jejuni, strain MOLcc20, 99% identity), MF872610 (Campylobacter jejuni, strain MOLcc29, 99% identity), MF872611 (Campylobacter jejuni, strain MOLcc30, 99% identity) and MF872612 (Lactobacillus reuteri, strain MOLcl43, 99% identity).
PCR detection of virulence gene determinants of Campylobacter species
To identify the virulence determinants of Campylobacter species isolated from chicken cloacal samples, four genes (cdtA, cdtB, cdtC and cadF) were targeted. The cdt gene is responsible for the expression of the cytolethal distension toxin which induces cell cycle arrest and promotes DNA damage.34,35 Consequently, its presence is associated with the severity of Campylobacter disease in humans. Figure 3 shows the amplification of the virulence genes of C. jejuni. In this study, the cdtB gene was detected in all (24, 100%) Campylobacter isolates while cdtA and cdtC genes were detected in 83.3% and 75% of the isolates, respectively. These results concur with the findings of studies by Gonzalez-Hein et al.36, Carvalho et al.37 and Casabonne et al.38 who detected cdtB genes in all Campylobacter species isolated from various sources. In contrast, Montwedi and Ateba25 reported relatively lower detection rates of cdtA (18/50, 36%) and cdtC (9/50, 18%) genes from chicken meat samples. The cadF gene encodes a protein that interacts with the host's fibronectin, consequently playing an important role in the adhesion and colonisation of the bacterium in the host's gut. In this study, the cadF gene was detected in the majority (18, 85.7%) of C. jejuni isolates, in line with previous studies which revealed close to 100% detection of the cadF gene.36,38,39 However, none of the C. coli isolates from the present study showed the presence of the cadF gene, possibly because only three C. coli isolates were tested. Despite the importance of the cadF gene, other genes such as the flaA and ciaB genes (not investigated in this study) are equally important in the adhesion, colonisation and, consequently, survival of bacteria in the gut.38
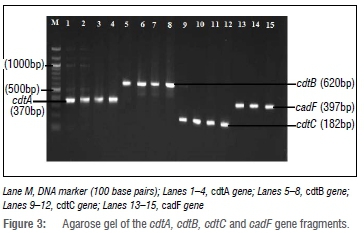
Genotyping of Campylobacter species
Genotyping was based on the combination of various identification and virulence genes. A total of eight genotypes designated G1 to G8 were detected among Campylobacter isolates (Table 2). G1 was the most prevalent (17, 71.4%) genotype among all Campylobacter species. Two genotypes (G1 and G7) possessed all three genes (cdtA, cdtB and cdtC) for toxin expression. These genotypes make up the majority of the Campylobacter isolated in this study. The CDT is a tripartite toxin with CDTB being the enzymatically active subunit; however, all three gene products are required for the toxin to be functionally active.34 Consequently, the majority of Campylobacter isolated in this study have the potential for producing functionally active toxins. As a result, we envisage a study to investigate the actual expression of toxins by Campylobacter isolates from environmental samples. In addition, most of the Campylobacter isolates as represented by G1, G3 and G5 possessed both genes for toxin production and adhesion. Interestingly, all the four C. jejuni strains that were sequenced belong to G1. The high prevalence of the cdt and cadF genes in Campylobacter species in this study is an indication of the pathogenic potential of the isolates.
Conclusion
Thermophilic campylobacters were successfully isolated from chicken cloacal swabs and the gene sequences of isolates deposited in GenBank. Highly virulent Campylobacter species were detected in chicken faeces in the study area, which may present a serious health hazard. Consequently, there is a need to implement control measures to curb the spread of these virulent strains.
Acknowledgements
We are grateful to the North-West University Postdoctoral Fellowship Programme for providing funds for this study and to the Department of Microbiology, North-West University (Mafikeng Campus) for providing facilities to conduct the study. We also acknowledge the assistance of the workers at Molelwane Farm in sample collection.
Authors' contributions
M.E.A.B.: Design of methodology, sample collection and laboratory analysis, data validation, writing and revision of the manuscript. C.N.A.: Conception and design of the study, data validation, revision and final approval of the manuscript.
References
1.Ryan KJ, Ray CG. Sherris medical microbiology: An introduction to infectious diseases. 4th ed. New York: McGraw Hill; 2004. p. 378-380. [ Links ]
2.Linscott AJ. Food-borne illnesses. Clin Microbiol Newsl. 2011;33(6):41-45. [ Links ]
3.Azevedo I, Albano H, Silva J, Teixeira P. Food safety in the domestic environment. Food Control. 2014;37:272-276. https://doi.org/10.1016/j.foodcont.2013.09.058 [ Links ]
4.Meunier M, Guyard-Nicodeme M, Dory D, Chemaly M. Control strategies against Campylobacter at the poultry production level: Biosecurity measures, feed additives and vaccination. J Appl Microbiol. 2015;120:1139-1173. https://doi.org/10.1111/jam.12986 [ Links ]
5.Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: A food production perspective. Int J Food Microbiol. 2007;117(3):237-257. https://doi.org/10.1016/j.ijfoodmicro.2007.01.006 [ Links ]
6.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, Teixeira P. Campylobacter spp as a foodborne pathogen: A review. Front Microbiol. 2011;2:200. https://doi.org/10.3389/fmicb.2011.00200 [ Links ]
7.Konkel ME, Monteville MR, Rivera-Amill V, Joens LA. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr Issues Intest Microbiol. 2001;2(2):55-71. [ Links ]
8.Rao MR, Naficy AB, Savarino SJ, Abu-Elyazeed R, Wierzba TF, Peruski LF, et al. Pathogenicity and convalescent excretion of Campylobacter in rural Egyptian children. Am J Epidemiol. 2001;154:166-173. https://doi.org/10.1093/aje/154.2.166 [ Links ]
9.Nyati KK, Nyati R. Role of Campylobacter jejuni infection in the pathogenesis of Guillain-Barre Syndrome: An update. BioMed Res Int. 2013;2013, Art. ID 852195, 13 pages. https://doi.org/10.1155/2013/852195 [ Links ]
10.Quinn PJ, Markey BK, Carter ME, Demnelly WJ, Leonard FC. Veterinary microbiology and microbial disease. 8th ed. Oxford: Blackwell Publishing; 2001. [ Links ]
11.Guerry P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007;15:456-461. https://doi.org/10.1016/j.tim.2007.09.006 [ Links ]
12.Grant CCR, Michael EK, Witold C Jr, Lucy ST. Role of flagella in adherence, internalization and translocation of Campylobacter jejuni in non-polarised and polarised epithelial cell cultures. Infect Immun. 1993;61:1764-1771. [ Links ]
13.Konkel ME, Garvis SG, Tipton SL, Anderson Jr DE, Cieplak Jr W. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953-963. https://doi.org/10.1046/j.1365-2958.1997.4031771.x [ Links ]
14.Lee RB, Hassane DC, Cottle DL, Pickett CL. Interactions of Campylobacter jejuni cytolethal distending toxin subunits CdtA and CdtC with HeLa cells. Infect Immun. 2003;71:4883-4890. https://doi.org/10.1128/iai.71.9.4883-4890.2003 [ Links ]
15.Lara-Tejero M, Galán JE. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect Immun. 2001;69(7):4358-4365. https://doi.org/10.1128/iai.69.7.4358-4365.2001 [ Links ]
16.US Centers for Disease Control and Prevention (CDC). Surveillance for foodborne disease outbreaks, United States, 2014: Annual report. Atlanta, GA: US Department of Health and Human Services, CDC; 2016. [ Links ]
17.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. Campylobacter jejuni - an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28-35. https://doi.org/10.3201/eid0501.990104 [ Links ]
18.Platts-Mills JA, Kosek M. Update on the burden of Campylobacter in developing countries. Curr Opin Infect Dis. 2014;27(5):444-450. https://doi.org/10.1097/qco.0000000000000091 [ Links ]
19.Mason J, Iturriza-Gomara M, O'Brien SJ, Ngwira BM, Dove W, Maiden MC, et al. Campylobacter infection in children in Malawi is common and is frequently associated with enteric virus co-infections. PLoS ONE. 2013;8, e59663, 5 pages. https://doi.org/10.1371/journal.pone.0059663 [ Links ]
20.Koulla-Shiro S, Loe C, Ekoe T. Prevalence of Campylobacter enteritis in children from Yaounde (Cameroon). Cent Afr J Med. 1995;41:91-94. [ Links ]
21.Lindblom GB, Ahren C, Changalucha J, Gabone R, Kaijser B, Nilsson LA, et al. Campylobacter jejuni/coli and enterotoxigenic Escherichia coli (ETEC) in faeces from children and adults in Tanzania. Scand J Infect Dis. 1995;27:589-593. https://doi.org/10.3109/00365549509047073 [ Links ]
22.Diergaardt SM, Venter SN, Spreeth A, Theron J, Brozel VS. The occurrence of campylobacters in water sources in South Africa. Water Res. 2004;38:2589-2595. https://doi.org/10.1016/j.watres.2004.03.004 [ Links ]
23.Samie A, Obi CL, Barret LJ, Powell SM, Guerrant RL. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool sample from the Venda region, Limpopo, South Africa: Studies using molecular diagnostic methods. J Infect. 2007;54(6):558-566. https://doi.org/10.1016/j.jinf.2006.10.047 [ Links ]
24.Mabote KI, Mbewe M, Ateba CN. Prevalence of Campylobacter contamination in fresh chicken meat and milk obtained from markets in the North-West Province, South Africa. J Hum Ecol. 2011;36(1):23-28. https://doi.org/10.1080/09709274.2011.11906413 [ Links ]
25.Montwedi MG, Ateba CN. Use of the cdt gene specific PCR in determining virulence properties of Campylobacter jejuni isolated from chicken meat samples obtained in some supermarkets in Mafikeng, NWP, South Africa. Life Sci J. 2012;9(3):2696-2701. [ Links ]
26.Prescott H. Laboratory exercise in microbiology. 5th ed. New York: McGraw Hill; 2002. [ Links ]
27.Persson S, Olsen KE. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol. 2005;54(11):1043-1047. https://doi.org/10.1099/jmm.0.46203-0 [ Links ]
28.Linton D, Lawson AJ, Owen RJ, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568-2572. [ Links ]
29.Frank JA, Reich CI, Wilson BA, Sharma S, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461-2470. https://doi.org/10.1128/aem.02272-07 [ Links ]
30.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003;52:345-348. https://doi.org/10.1099/jmm.0.05056-0 [ Links ]
31.Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999;38:510-517. [ Links ]
32.Uaboi-Egbenni PO, Bessong PO, Samie A, Obi CL. Prevalence and antimicrobial susceptibility profiles of Campylobacter jejuni and coli isolated from diarrheic and non-diarrheic goat faeces in Venda region, South Africa. Afr J Biotechnol. 2011;10(64):14116-14124. https://doi.org/10.5897/ajb10.1662 [ Links ]
33.Kumar PA, Kumar S. Comparative analysis of cultural and PCR based assays for detection of Campylobacter spp. in human stool samples. Proc Natl Acad Sci India. 2015;85(3):839-844. https://doi.org/10.1007/s40011-015-0565-2 [ Links ]
34.Asakura M, Samosornsuk W, Hinenoya A, Misawa N, Nishimura K, Matsuhisa A, et al. Development of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the detection and identification of Campylobacter jejuni, Campylobacter coli and Campylobacter fetus. FEMS Immunol Med Microbiol. 2008;52:260-266. https://doi.org/10.1111/j.1574-695x.2007.00369.x [ Links ]
35.Bolton DJ. Campylobacter virulence and survival factors. Food Microbiol. 2015;48:99-108. [ Links ]
36.González-Hein G, Huaracán B, Garcia P, Figueroa G. Prevalence of virulence genes in strains of Campylobacter jejuni isolated from human, bovine and broiler. Braz J Microbiol. 2013;44(4):1223-1229. https://doi.org/10.1590/s1517-83822013000400028 [ Links ]
37.Carvalho AF, Silva DM, Azevedo SS, Piatti RM, Genovez ME, Scarcelli E. Detection of CDT toxin genes in Campylobacter spp. strains isolated from broiler carcasses and vegetables in São Paulo, Brazil. Braz J Microbiol. 2013;44(3):693-699. https://doi.org/10.1590/s1517-83822013000300005 [ Links ]
38.Casabonne C, Gonzalez A, Aquili V, Subils T, Balague C. Prevalence of seven virulence genes of Campylobacter jejuni isolated from patients with diarrhea in Rosario, Argentina. Int J Infect. 2016;3(4), e37727, 6 pages. https://doi.org/10.17795/iji-37727 [ Links ]
39.Zhang T, Luo Q, Chen Y, Li T, Wen G, Zhang R, et al. Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 2016;8, Art. #48, 9 pages. https://doi.org/10.1186/s13099-016-0132-2 [ Links ]
 Correspondence:
Correspondence:
Marie Bissong
mabissong@yahoo.com
Received: 19 Mar. 2018
Revised: 26 Oct. 2018
Accepted: 02 July 2019
Published: 26 Sep. 2019
EDITOR: John Butler-Adam
FUNDING: North-West University
Supplementary Material
The open data set is available here: [Open data set]














