Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.115 no.3-4 Pretoria mar./abr. 2019
http://dx.doi.org/10.17159/sajs.2019/5482
COMMENTARIES
Managing South African biodiversity research data: Meeting the challenges of rapidly developing information technology
Willem CoetzerI; Michelle HamerII, III
ISouth African Institute for Aquatic Biodiversity, Grahamstown, South Africa
IISouth African National Biodiversity Institute, Pretoria, South Africa
IIISchool of Life Sciences, University of KwaZulu-Natal, Pietermaritzburg, South Africa
Keywords: natural science collections; biodiversity data curation; biodiversity occurrence data; data integration; capacity development
New developments in the funding requirements of biodiversity science as well as rapidly developing information technology warrant a sharper focus on the way in which biodiversity data are managed. We propose that an opportunity presents itself to develop a specific set of informatics skills among a new class of data analysts in the biodiversity science community. Our consideration of capacity development specifically emphasises the need for conceptual rigour, compliance with technical data standards and the culture of data publication or data sharing.
There is a pressing need for data stewardship skills and positions in the South African biodiversity science community. We describe previous and current initiatives that may help to provide the context of, and develop skills and capacity for, effective management or stewardship of biodiversity research data. The overlapping competencies of data stewardship, data curation and data preservation include:
processes and activities related to the organization and integration of data collected from various sources, annotation of the data, and publication and presentation of the data such that the value of the data is maintained over time, and the data remains available for reuse and preservation.1
The role of the data steward can be distilled into the fundamental principles of findability, accessibility, interoperability and reusability (the FAIR principles).1
From where do biodiversity data originate?
During the last 5 years, a significant component of funding for South African biodiversity science has been channelled through the Foundational Biodiversity Information Programme (FBIP).2 The FBIP is funded by the South African Department of Science and Technology (DST) and administered by the National Research Foundation and the South African National Biodiversity Institute (SANBI). The FBIP recognises the importance of biodiversity, not only in the narrower sense of a particular discipline of scientific research (e.g. taxonomy, systematics or ecology), but also in the broader context of the relevance of biodiversity to society. Four large, collaborative FBIP projects have been funded. These projects focus on marine biodiversity (the Seakeys Project), the effect of habitat fragmentation on the faunal diversity of Eastern Cape forests, filling gaps in biodiversity information to support decisions about the exploitation of shale gas in the Karoo (the Biogaps Project), and camera trapping of mammals to assess the status of species and populations inside and outside protected areas (the Snapshot Safari Project). In 2016, 20 smaller FBIP projects were undertaken to investigate a variety of subjects, including bat monitoring in the Kruger National Park, bryozoan e-taxonomy, and a number of applied projects, e.g. the use of polychaetes as bait, and a survey of earthworms and their use in vermicomposting. The FBIP explicitly requires researchers to generate and submit research data characterised as species occurrences, species attributes or population abundance records, or develop tools or generate data that facilitate the identification of species, including through molecular techniques (e.g. taxonomic keys or DNA barcodes). Physical specimens may or may not be preserved in the execution of these research projects. Resultant occurrence records may be associated with high-quality still images, videos or sound recordings.
The community of natural history collections (more appropriately referred to as natural science collections, or NSC) naturally intersects with the community of biodiversity researchers funded by the FBIP. Recent developments among South African NSC museums, including increased funding, promise to improve the conditions, operations and utilisation of South African NSC (see below). Much has been written about the use of NSC or NSC data.3,4 Such uses include estimating the spread of invasive alien or pest species; evaluating the abundance, conservation status and distribution of threatened species5-7; or projecting the ecosystem impacts of urban development, e.g. changes in ecosystem services such as pollination8.
Properly and efficiently managing the biodiversity research data described above presents technical and organisational challenges arising from the rapid development of technology. Researchers' or technicians' data management skills do not always match the increasingly stringent requirements to organise and store data from the broad and diverse array of biodiversity projects conducted by the South African research community. These requirements are both technical and administrative (e.g. that data should be available for others to use). How can we improve biodiversity data management, integration and utilisation (e.g. how should students collaborate with their supervisors to share data, especially when they are not on the same campus)? Where should the data be stored and who should be responsible for data storage and long-term data preservation? Which data standards should be used? What conditions should be associated with using the data? Below we describe some of the challenges that the broader community of biodiversity scientists could face in developing greater capacity, specifically to manage and meaningfully use biodiversity occurrence data.
The need for conceptual rigour in curating NSC or biodiversity data
Collectively, records of physical specimens and records of observations of organisms are termed 'occurrence records' - hence we speak about 'the occurrence of a species at a place and time'. This phrase encapsulates all the fundamental classes of knowledge (i.e. metadata) about most of the biodiversity data referred to above. Occurrence records are particularly important to anchor abstract knowledge of species in the observed world. For example, an ecologist may need to assess the (occurrence of) freshwater invertebrate indicator species in a particular stream to evaluate its current state, or compare the arthropod community structure of a forest with that of a nearby crop to assess the availability of natural enemies.
All such biodiversity occurrence records need to be curated in a specifically designed biodiversity database, even if representative voucher specimens are not preserved and deposited in a natural science museum for future reference. Occurrences of certain species may not be found, and these absences can be meaningful, e.g. when plants or marine invertebrates are systematically sampled using quadrats or photo-quadrats. Records of systematically structured sampling events and transects are therefore important to know that any effort, or how much comparable effort, was made to find occurrences. Species' absences increase the rigour of analyses such as ecological niche modelling, in which the species distribution range is estimated.
To comprehensively characterise the context of data, sampling events and occurrences must be represented using a coherent conceptual model. At a higher level of conceptual abstraction in this model, physical specimens and human observations are represented by the same properties of metadata classes. After all, a bird (occurrence) may have been seen during a sampling event at a particular place, whether or not the bird was captured or preserved as a specimen (Figure 1).
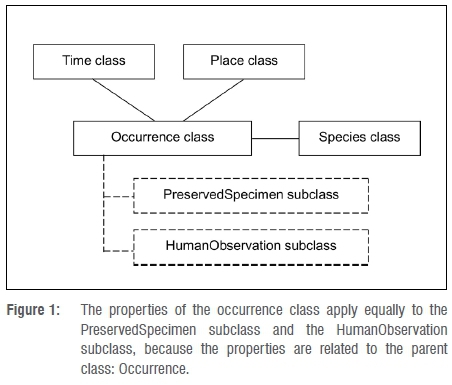
There is thus a need to develop skills and capacity for generalised biodiversity data curation or stewardship, to integrate data records representing the full suite of concepts used by scientists, or to integrate typical NSC data with typical ecological data (i.e. to integrate specimen records with observations) for greater rigour or broader spatio-temporal coverage.
Below we elaborate on the basic idea, supported by the above reasoning, that a particular biodiversity database application (specifically its database schema), which is open-source software, is an ideal tool to use, both for physical specimens and biodiversity observations. In other words, it is an ideal database and application to manage biodiversity sampling event and occurrence records. Wider adoption of a common conceptual model, data management protocol, and approach will foster the development of a future class of biodiversity informatics technicians and analysts who will be able to efficiently manage and preserve our biodiversity research data.
Moving beyond traditional uses of NSC collection databases
The traditional specimen collection database is useful within the NSC museum, to document and manage a museum's specimen holdings by making inventories and keeping track of loans. More rigorous attention to the curation of biodiversity occurrence records will address other practical needs, e.g. the increasing requirements to include data management plans in funding proposals and upload data sets to stable, online repositories.
Current biodiversity database applications include fields and functions which serve purposes other than collection management. For example, a globally unique identifier (GUID) ensures that a record can be uniquely identified, i.e. not confused with any other record published on the World Wide Web, which can be seen as the 'extended database' that is used to publish or share data. Such web technologies (of which GUIDs are just one example) are indicative of the changing culture of scientific data use typical of the Open Science Movement. These technologies imply that researchers ought to publish their biodiversity data in a way that makes the standardised (meta)data accessible to other researchers (i.e. researchers ought to use this extended database properly) (see Box 1).9 The data steward therefore needs a thorough understanding of the conceptual model of the local database as well as that of the online repository or relevant data standard (see below).

The Open Science Movement3,10 offers many diverse motivations to share scientific data, publications and knowledge, and mechanisms for conducting open scientific research. In South Africa, a new multi-institutional initiative, the Data Intensive Research Initiative of South Africa (DIRISA), is aligned with the principles of open science. The first National Research Data Workshop was held from 19 to 21 June 2018, and included presentations from astronomers, sustainable development researchers, bioinformaticians, biodiversity scientists and librarians, among others.11 DIRISA is one of the three pillars of the National Integrated Cyber Infrastructure System (NICIS), an initiative of the DST that is implemented by the Council for Scientific and Industrial Research (CSIR). The other two pillars of the NICIS are the Centre for High Performance Computing (CHPC) and the South African National Research Network (SANReN).
The Global Biodiversity Information Facility (GBIF) currently publishes just over 1 billion standardised biodiversity occurrence records, from about 39 000 data sets contributed by about 1000 data providers around the world (Figure 2). Of these, 77% are records of human observations, and only 15% are records of physical specimens. Of the 19.2 million occurrences on the GBIF Data Portal originating from South Africa, only 1.8 million (9.4%) are preserved specimens. The GBIF data are freely available to be used in accordance with the terms of three Creative Commons (CC) licences. Many data providers will require attribution according to a supplied citation and will therefore publish their data under a CC-BY licence. Other data providers commit their data to the public domain and publish under a CC-Zero licence (not necessarily requiring acknowledgement or citation), or stipulate a CC-BY-NC licence, adding the requirement that use of the data will not be for commercial purposes. Occurrence records of southern African aquatic biodiversity are published on the GBIF Data Portal by the South African Institute for Aquatic Biodiversity (SAIAB) under a CC-BY licence.16
Improving biodiversity data curation in South African natural science collections
In 2012, the Museum Data Migration Project18 was initiated by SAIAB to migrate the specimen records of selected museums to newly developed collection databases. Museum staff were then trained to use the databases to better manage specimen collections. Specify Software19, which has been under development for about 30 years, was used to develop the databases. Specify Software is popular worldwide and is currently used by about 60 trained users working in 13 South African NSC museums (which house more than 50 specimen collections). From 2012 to 2015, another project, funded by the JRS Biodiversity Foundation, involved the cleaning and migration of significant data sets of arachnid and other data to new or existing collection databases, accompanied by further Specify Software training.
South African NSC have been periodically assessed since 1974.20 Despite recognition of their importance, globally NSC have not fared well because of decreasing funding and the erosion of positions.21 South Africa is a shining exception since the launch in October 2017 of the Natural Science Collections Facility (NSCF) - a much-anticipated response to concerns of neglected collections raised in recent years by the biodiversity research community. The NSCF is a virtual facility composed of a network of institutions that hold natural science collections which are accessible to external researchers. The overall aim of the NSCF is to ensure that natural science research collections and associated data are used for high-quality research and decision-making to address issues of socio-economic importance. The NSCF is funded as part of the DST's long-term funding programme, the Research Infrastructure Roadmap (SARIR), and administered by SANBI. A Coordinating Committee oversees operational management and is supported by several working groups made up of staff already employed by South African NSC museums. The Data Working Group includes representatives from various collection institutions who have experience in data management and strive to improve data curation and the use of appropriate data standards across institutions, to enable integration and publication of high-quality, standardised biodiversity data.
A new initiative, the Biodiversity Data Curation Platform (comparable to a cloud hosting service), initiated by SAIAB, will build on the Museum Data Migration Project by offering South African museums dedicated webservers and Specify 7 databases. Specify 7 is a web application that is the latest product released by the Specify Collections Consortium. It is hoped that the Biodiversity Data Curation Platform will ease museums' data management burden and contribute towards the objectives of the NSCF. Rather than requiring their own database server or systems administration expertise, staff of a participating NSC museum can gain access to a database on a virtual server, simply by loading a website using a standard web browser. Nothing else is required to make the museum's customised Specify 7 database and application available to perform routine collection management functions (e.g. catalogue specimens, query data, create loan records, and print loan invoices or specimen labels) or advanced informatics functions (e.g. export standardised data for publication on the GBIF Data Portal). In 2018, the vertebrate specimen records of four NSC museums, which have not previously used Specify Software for vertebrate specimens, were migrated to newly created databases hosted by the Biodiversity Data Curation Platform. Vertebrate specimens have been prioritised by the NSCF, both in terms of physical curation and data curation. These specimens and records are now in a better state to be examined by expert taxonomists, and brought to the requisite standard of preservation and information (e.g. specimens may need to be re-identified, and the taxonomy reflected in databases brought up-to-date). It is hoped that the Biodiversity Data Curation Platform will foster the development of biodiversity data curation expertise in South Africa's natural science museums.
Specify Software is not only for managing collections of physical specimens
An important SAIAB research project typifies the kind of biodiversity occurrence data in the South African community that need to be brought under formal data curation, namely the work on Baited Remote Underwater Video (or BRUV, another research platform offered by SAIAB)22, and the closely related work on marine macrobenthos imagery23. The data will inform better decisions about the management of reef ecosystems and fish resources. In this underwater camera-trap and photo-quadrat sampling work, collection of physical specimens is not among the objectives. The BRUV videos and still images (Figure 3) of subtidal reef fish and macrobenthos are associated with (meta)data that are used to assess fish assemblage structure, including species composition, abundance and size. The number of fish observed is recorded and the lengths of some of these are measured (if stereo cameras are used). Standard spatio-temporal metadata (place and time) as well as instrument settings are also recorded.

The data generated by this project are therefore typical human observations of biodiversity (marine fish; in the case of macrobenthos the data are typical photo-quadrat sampling events to estimate percentage cover, including species absences). The conceptual model and database schema underlying Specify Software was tested to evaluate whether any of the fields necessitated by the fish and macrobenthos data and metadata could be said to be excluded. It was found that all fields were easily accommodated by the database schema.
When Specify Software is re-used for biodiversity observation data, interaction with the data need not be limited to the use of the Specify Software interface, but can be achieved through a custom-developed user-form (Figure 4) specifically tailored to users' various requirements. In contrast, tailoring a database schema and input mechanism (by far the heavier infrastructure), or underlying conceptual model, to each biodiversity research project would be tantamount to re-inventing the wheel many times, and would complicate data integration.
We therefore argue that capacity development for the curation of biodiversity occurrence records, including many or most of the different biodiversity sampling protocols and objectives (i.e. not only traditional NSC objectives), can potentially be strengthened by the use of a common conceptual model (database schema) and related 'spoken language'. By re-using the Specify database schema we will be standardising the tools we use to carry out the same fundamental operations of information management across the community (e.g. data validation preceding batch data importation), which will make it easier for technicians to learn the techniques of biodiversity data curation.
The Biodiversity Data Curation Platform includes a tool to publish data, but the platform allows the NSC museum clients to execute information management functions independently and according to their own procedures and policies. The Integrated Publishing Toolkit (IPT) was developed by GBIF to simplify the process of publishing standardised biodiversity data on the GBIF Data Portal. At the national scale, fish and invertebrate sampling protocols may be differently designed and metadata classes differently defined from project to project, and this could complicate the storage, management, sharing, analysis and interpretation of data. Use of the Biodiversity Data Curation Platform could therefore be a first step to remedy this semantic heterogeneity, by allowing different users to manage their data independently but in a way that will allow the data to be vertically integrated (Figure 5) through the use of biodiversity information standards (specifically the Darwin Core metadata terms24).
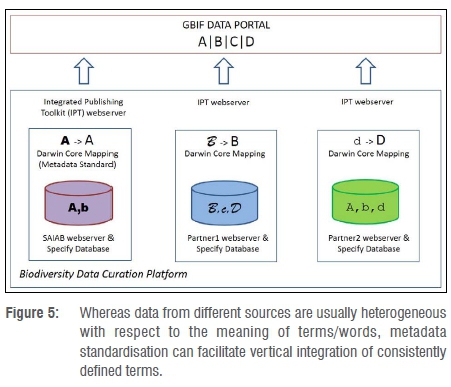
When publishing biodiversity or occurrence data it is important to use ratified data standards. Compliance with these standards basically requires particular words to be used as field names (e.g. 'basisOfRecord') provided that a strict definition applies, as well as particular words to be used as the data values (e.g. 'HumanObservation') in these standardised fields. Biodiversity data standards are developed and published by the community of biodiversity informatics practitioners and researchers, through the organisation Biodiversity Information Standards (formerly the Taxonomic Databases Working Group).25
Challenges and capacity development
We need to investigate ways to further develop technical skills to use Specify 7 technology effectively in NSC museums and biodiversity research institutes. Ensuring that the transition to Specify Software will be sustainable must be a high priority. It will be important to design a comprehensive training programme to improve data management, data curation and data publication skills in the NSC and biodiversity science community. Only then can we expect that the increasing use of information technology in NSC and associated institutes will become differentiated into new roles in these organisations. It is possibly this lack of differentiation that has held back the development of biodiversity informatics skills and professionals.
The process of 'cleaning' legacy NSC and biodiversity data, and migrating the data to a new, more rigorous database schema, is potentially a bottleneck to progress. Even when all the legacy data have been migrated to the new platform, a national-scale, sustained effort will be required to ensure that newly acquired data will continue to be imported into, and curated in, NSC databases consistently, timeously and accurately. The Specify suite of applications includes a 'Workbench' tool which can be used to map spreadsheet columns to database fields, for bulk data importation. The outcome of a given data importation routine will depend on the data steward's understanding of how the conceptual model represents knowledge concepts denoted by the (meta)data. This is therefore where the focus of capacity development for biodiversity data curation should be (i.e. rather than focusing simply on using the Specify application's interface correctly to catalogue individual records, which requires a much lower level of competence). The Specify Workbench is an important tool and level of technology development, around which concrete capacity development initiatives can be designed, to engage not only specialist data analysts but traditional NSC museum practitioners as well.
This new class of data stewards will be responsible for carefully channelling the flow of data into NSC or biodiversity databases and sharing the data for wider use. Enhancing data curation skills could contribute to the establishment of a new culture of data stewardship in NSC and biodiversity research institutes, in which South African biodiversity researchers and technicians can look forward to collaborating on exciting and creative projects to use new information technology and high-quality data in biodiversity science and ecological research.
References
1.Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3, Art. #160018, 9 pages. Available from: https://doi.org/10.1038/sdata.2016.18 [ Links ]
2.Foundational Biodiversity Information Programme [homepage on the Internet]. No date [cited 2018 Jul 01]. Available from: http://fbip.co.za/ [ Links ]
3.Schilthuizen M, Vairappan CS, Slade EM, Mann DJ, Miller JA. Specimens as primary data: Museums and "open science". Trends Ecol Evol. 2015;30:237-238. Available from: https://doi.org/10.1016/j.tree.2015.03.002 [ Links ]
4.Coetzer W. Natural science collections [network on the Internet]. c2018 [cited 2018 Jul 01]. Available from: https://www.mendeley.com/community/natural-science-collections/ [ Links ]
5.Ponder WF, Carter GA, Flemons P, Chapman RR. Evaluation of museum collection data for use in biodiversity assessment. Conserv Biol. 2001;15(3):648-657. Available from: http://doi.wiley.com/10.1046/j.1523-1739.2001.015003648.x [ Links ]
6.Drinkrow DR, Cherry MI, Siegfried WR. The role of natural history museums in preserving biodiversity in South Africa. S Afr J Sci. 1994;90(8-9):470-479. [ Links ]
7.Tweddle D, Bills R, Swartz E, Coetzer W, Da Costa L, Engelbrecht J, et al. The status and distribution of freshwater fishes. In: Darwall WRT, Smith KG, Tweddle D, Skelton P, editors. The status and distribution of freshwater biodiversity in southern Africa. Gland/Grahamstown: IUCN/South African Institute for Aquatic Biodiversity; 2009. p. 21-37. [ Links ]
8.Pauw A, Hawkins JA. Reconstruction of historical pollination rates reveals linked declines of pollinators and plants. Oikos. 2011;120(3):344-349. Available from: http://centaur.reading.ac.uk/20920/ [ Links ]
9.Costello MJ, Michener WK, Gahegan M, Zhang Z-Q, Bourne PE. Biodiversity data should be published, cited, and peer reviewed. Trends Ecol Evol. 2013;28(8):454-461. https://doi.org/10.1016/j.tree.2013.05.002 [ Links ]
10.Lin T. Cracking open the scientific process. New York Times. 2012 January 16. Available from: https://www.nytimes.com/2012/01/17/science/open-science-challenges-journal-tradition-with-web-collaboration.html?_r=2 [ Links ]
11.DIRISA. National Research Data Workshop [webpage on the Internet]. c2018 [cited 2018 Jul 07]. Available from: https://www.dirisa.ac.za/workshop/ [ Links ]
12.Chavan V, Penev L. The data paper: A mechanism to incentivize data publishing in biodiversity science. BMC Bioinformatics. 2011;12(Suppl 15), S2, 12 pages. Available from: https://doi.org/10.1186/1471-2105-12-S15-S2 [ Links ]
13.Koopman MM, De Jager K. Archiving South African digital research data: How ready are we? S Afr J Sci. 2016;112(7/8), Art. #2015-0316, 7 pages. https://doi.org/10.17159/sajs.2016/20150316 [ Links ]
14.Tenopir C, Allard S, Douglass K, Aydinoglu AU, Wu L, Read E, et al. Data sharing by scientists: Practices and perceptions. PLoS ONE. 2011;6(6), e21101, 21 pages. https://doi.org/10.1371/journal.pone.0021101 [ Links ]
15.Penev L, Mietchen D, Chavan V, Hagedorn G, Smith V, Shotton D, et al. Strategies and guidelines for scholarly publishing of biodiversity data. Res Ideas Outcomes. 2017;3, e12431, 12 pages. https://doi.org/10.3897/rio.3.e12431 [ Links ]
16.Coetzer W. Occurrence records of southern African aquatic biodiversity [data set on the Internet]. c2010 [cited 2018 Jul 06]. Available from: https://www.gbif.org/dataset/1aaec653-c71c-4695-9b6e-0e26214dd817 [ Links ]
17.Open Streetmap. Copyright and license [webpage on the Internet]. No date [cited 2019 Feb 13]. Available from: http://www.openstreetmap.org/copyright [ Links ]
18.Coetzer W, Gon O, Hamer M, Parker-Allie F. A new era for specimen databases and biodiversity information management in South Africa. Biodivers Informatics. 2012;8:1-11. https://doi.org/10.17161/bi.v8i1.4263%09%0A [ Links ]
19.Specify Software [software on the Internet]. No date [cited 2018 Feb 01]. Available from: http://www.specifysoftware.org [ Links ]
20.Coaton W. Status of the taxonomy of the Hexapoda of southern Africa. Entomol Mem Dep Agric Tech Serv Repub South Africa. 1974;38:1-124. [ Links ]
21.Kemp C. Museums: The endangered dead. Nature. 2015;518(7539):292-294. https://doi.org/10.1038/518292a [ Links ]
22.Bernard ATF, Götz A, Parker D, Heyns ER, Halse SJ, Riddin NA, et al. New possibilities for research on reef fish across the continental shelf of South Africa. S Afr J Sci. 2014;110(9/10), #a0079, 5 pages. http://dx.doi.org/10.1590/sajs.2014/a0079 [ Links ]
23.Heyns ER, Bernard ATF, Richoux NB, Götz A. Depth-related distribution patterns of subtidal macrobenthos in a well-established marine protected area. Mar Biol. 2016;163(2):1-15. [ Links ]
24.Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, et al. Darwin Core: An evolving community-developed biodiversity data standard. PLoS ONE. 2012;7(1), e29715, 8 pages. Available from: https://doi.org/10.1371/journal.pone.0029715 [ Links ]
25.Biodiversity Information Standards [homepage on the Internet]. No date [cited 2018 Feb 01]. Available from: http://www.tdwg.org/ [ Links ]
 Correspondence:
Correspondence:
Willem Coetzer
W.Coetzer@saiab.ac.za
Published: 27 March 2019














