Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.114 no.9-10 Pretoria sep./oct. 2018
http://dx.doi.org/10.17159/sajs.2018/4508
REVIEW ARTICLE
Emergence of vancomycin-resistant enterococci in South Africa: Implications for public health
Frank E. Tatsing Foka; Ajay Kumar; Collins N. Ateba
Department of Microbiology, School of Biological Sciences, Faculty of Natural and Agricultural Sciences, North-West University, Mafikeng, South Africa
ABSTRACT
South Africa is among the countries with the highest prevalence of debilitating diseases such as HIV/Aids and diabetes. In this context, the emergence of vancomycin-resistant enterococci (VREs) in most South African ecological niches is quite disturbing, taking into consideration the fact that therapeutic options in a case of resistant-enterococci infection would be limited. Agricultural practices coupled with the misuse of antibiotics in intensive animal rearing and in hospital facilities have led to the creation of reservoirs of VREs in the environment. VREs can cause serious health problems by transmitting their resistance genes to susceptible pathogens; they are transmitted to humans by direct or indirect contact and through the food chain. We screened thoroughly the AJOL and the PubMed databases for studies on VRE incidence in South Africa. This review gives insight into the current status of antimicrobial resistance management in South Africa; it explores the different pathways involved in the spread of VREs and proposes possible solutions to tackle the issue of VREs and antimicrobial resistance in South Africa and other parts of the world.
Significance:
• The recent detection of vancomycin-resistant enterococci in most South African ecological niches poses a serious threat to public health and is therefore an issue of great concern.
• This study not only addresses the causes and patterns of resistance to antimicrobial agents, particularly in South Africa, but also outlines a holistic approach to potential strategies to tackle antimicrobial resistance in South Africa and the world at large.
Keywords: antimicrobial resistance; glycopeptide resistance; emerging pathogens
Introduction
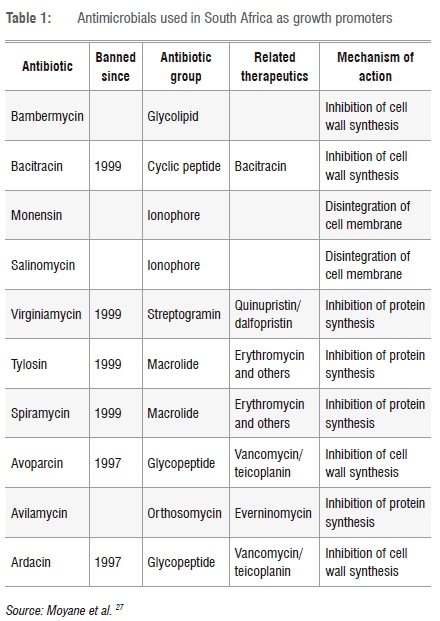
Shortly after antibiotics were introduced for therapeutic purposes, their growth-promoting attributes were discovered; ever since, most antibiotics and their analogues have been used as growth promoters in animal farming.1 Growth promoters are believed to improve feed conversion, promote animal growth and reduce mortality and morbidity rates resulting from clinical and subclinical illnesses, although the mechanisms through which these effects are achieved are still poorly understood.2 This effect of antibiotics motivated the use of avoparcin as a growth promoter for many decades before it was banned worldwide as a result of the emergence of vancomycin-resistant enterococci (VREs).3 The rise of VREs reduced the efficacy of enterococcal infection treatments with teicoplanin and vancomycin (which were the drugs of choice until then), making treatment more challenging.
The isolation of VREs in hospitals and environmental samples worldwide and specifically in South Africa is a serious health concern.4-6 In fact, VREs were isolated from surface water in the Netherlands, from ground water and from hospital waste water in South Africa.4-7 VREs mainly cause illnesses in immunocompromised hosts and in patients who are admitted to intensive care units for lengthy periods.8 For instance, VREs are responsible for endocarditis, urinary tract infections, bacteraemia, intra-abdominal and pelvic infections, and burn wound and deep tissue infections.
Because of the serious implications of VREs on public health, the issue of VREs cannot be underestimated. This review is therefore not just a citation of reports of VREs in South Africa and their patterns of spread and dissemination, but also an insight into the current management of antimicrobial resistance issues in South Africa with some recommendations on how to tackle the issue of vancomycin resistance genes in South Africa and other countries.
Mechanism of resistance to vancomycin
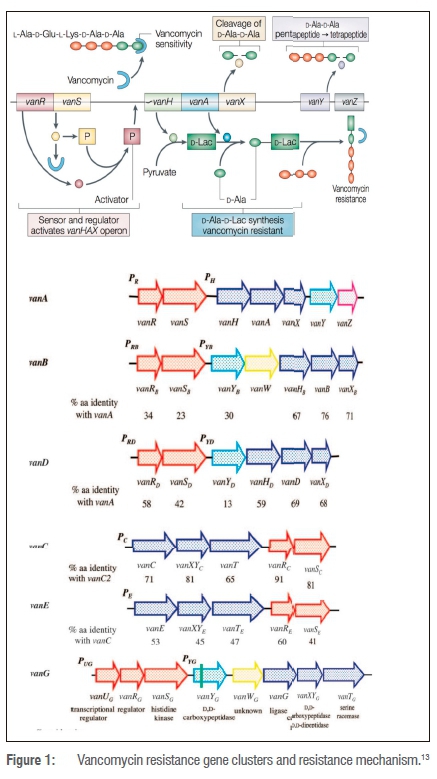
Two types of vancomycin resistance in enterococci have been reported so far: intrinsic and acquired resistance. Intrinsic resistance refers to an antimicrobial drug being ineffective as a result of inherent features in a species, like restricting drug accessibility to the target or not having the drug target. Acquired resistance occurs when the bacterium is initially susceptible, but develops resistance either by somatic mutation or by acquisition of genes by horizontal transfer. Characterised by low-level resistance to vancomycin, intrinsic resistance is commonly detected in Enterococcus gallinarum, E. casseliflavus and E. flavescens. As opposed to E. faecium and E. faecalis and less often E. raffinosus, E. avium and E. durans display acquired resistance to vancomycin resulting from the acquisition of genetic determinants either from another organism or from the environment.9-11 Resistance to vancomycin is conferred by 10 gene clusters: vanA, vanB, vanC, vanD, vanE, vanF, vanG, vanL, vanM and vanN. Gene clusters vanA, vanB, vanC, vanD, vanE and vanG have been extensively studied (Figure 1). The resistance phenotypes share the same basic mechanism of resistance. The glycopeptides bind to the carboxy-terminal D-Ala residues of cell wall precursors, thus preventing their incorporation into the nascent peptidoglycan. Substituting the terminal D-Ala residue with either D-lactate (vanA, vanB and vanD genotypes) or D-serine (vanC and vanE genotypes) confers resistance (Figure 1).11,12
Studies of vancomycin-resistant enterococci in South Africa
Vancomycin-resistant enterococci in food items and the environment
Very few reviews actually give an account of the situation in South Africa with respect to VREs in food items and the environment. Enterococci were isolated from lettuce and spinach leaves in the North West Province using polymerase chain reaction techniques.14 That study was motivated by the fact that enterococci had previously been screened from ground water intended for drinking in the same area.5,6 Vegetables were contaminated as a result of their proximity to the soil when they were grown.15 Animals and birds were another possible source of contamination but the water used in the irrigation process of these vegetables was a source of contamination. Moreover, the harvesting, packaging, handling and retail hygiene practices were other sources of contamination.15 As similar findings were made in Oman16, detection of VREs in fresh, leafy food items constitutes a serious health issue because lettuce is mostly eaten raw, in the form of salads.
Vancomycin-resistant enterococci were isolated from ground water intended for drinking in rural communities of the North West Province.5 The investigations indicated that limited access to proper sanitary facilities and lack of hygiene were the causative factors because the water sources were contaminated by faecal matter. These factors are the same as those that contributed to the contamination by VREs of municipal tap water in Mafikeng households, also in the North West Province.17 In addition to these causes, contamination of environmental soil and water bodies by resistant isolates can arise as a result of farming activities and contamination from other sources such as contaminated waste effluents from clinical settings.4,18,19
Vancomycin-resistant enterococci in hospital settings
Vancomycin-resistant enterococci constitute a serious issue in South African hospitals, especially taking into consideration the high incidence of HIV-positive patients and the high prevalence of other debilitating illnesses such as tuberculosis and diabetes. A possible synergistic association between Aids or any of the above-mentioned illnesses and VRE infections is rather unsettling and dreaded. In fact, clinical isolates of VREs were reported in South Africa for the first time in 1997.20 A few years later, a case report of VRE infections was filed at the paediatric oncology ward of a tertiary-level paediatric hospital in Cape Town and in the haematology unit of a similar type of hospital in Durban.19-22 Based on the results obtained from gene-sequencing assays (PFGE and MLST), it was suggested that a possible transfer of VRE isolates between patients or persistence of this isolate within the haematology/oncology unit were responsible.19 This resistance resulted from previous treatments with broad-spectrum antibiotics (mostly third-generation cephalosporins and carbapenems), prolonged stays in the hospital setting, vancomycin exposure, immunosuppression therapy and young age in the case of children.19-22 Moreover, in a recent study, the presence of VREs was reported in wastewater effluents from a hospital facility in Alice, Eastern Cape Province.4 These findings highlight the poor or inefficient water-treatment system of that facility's effluents before discharge into the environment. Antibiotics do not undergo total biodegradation in wastewater management processes; this hospital waste water was therefore regarded as a reservoir of resistant pathogens and an ideal environment for exchange of resistance genes or transfer to non-resistant isolates.23-25 These findings therefore demonstrate that the Victoria hospital waste water was a significant source of VREs in the wastewater treatment plant of Fort Hare. As VREs were screened from cow dung and environmental water sources in three selected dairy farms in the Amathole District, waste water from Victoria hospital must have a huge impact on the microbiological quality of the neighbouring environmental water bodies.26
Vancomycin-resistant enterococci in farming and agricultural practices
The Eastern Cape Province and part of the North West Province are mostly agrarian areas with countless numbers of piggery, poultry and cattle farms. Antimicrobials are used for the enhancement of productivity in South Africa.27,28 The shedding of resistant bacteria in the environment through faecal contamination is a concern. VREs that possessed vanB and vanC1/C2/C3 resistance genes were screened in pig dung in the Eastern Cape.28 Antibiotic resistance genes were spread in the environment as a result of excretion, flushing of out-of-date prescriptions, medical waste, leakage of septic tanks, effluents from wastewater treatment plants and agricultural waste discharges.28 The subtherapeutic doses of antimicrobials added in the animal feeds for prophylactic purposes and growth-promoting effects were the major causes of the resistance observed in the isolates.29,30 In fact, growth promoters have been associated worldwide with the rise of resistant bacteria, leading to the banning of most growth promoters.1-3,27
Pathways of antimicrobial resistance transmission
Antimicrobial agents are not fully transformed into inactive compounds in the systems of treated animals and are excreted in manure where they revert to their initial state after some time.31 This makes manure a hotspot for isolates carrying mobile genetic resistance elements and when mixed with soil for agricultural purposes, antibiotic resistance genes are likely to be vertically and horizontally transferred to soil bacteria. This transference leads to the pollution of soil by antibiotic resistance genes and aids in the uptake of antimicrobial resistance by commensal bacteria and human pathogens such as enterococci.32-34 Moreover, treated waste water is known to harbour antibiotic resistance genes which can be transferred to vegetables and crops through irrigation water35 (Figure 2).
There are several means through which resistant bacteria spread to humans, whether it be from food or from farm animals (Figure 2). The most plausible ones are via the food chain; direct or indirect contact with persons working in close proximity with animals such as farmers and animal healthcare professionals; and environmental components contaminated by agricultural waste or manure.2,32-38 Environment plays a key role as a potential reservoir of resistance genes whereby genetic determinants are exchanged amongst isolates that are taken up by humans and animals.4,39 Also, prolonged stay in a hospital setting can lead to patients acquiring VREs.4
Minimally processed food items, and raw and fermented food items constitute a potential risk through considerable numbers of viable cells; these food items could interact with other factors, such as cohabitation with pathogens, leading to the appearance of resistant strains of enterococci in the human gastrointestinal tract. The in-vitro transfer of erythromycin resistance genes from lactic acid bacteria to Listeria monocytogenes has been demonstrated.40-42 Moreover, numerous reports have highlighted the transfer of resistance genes amongst isolates of the same species in the human gut.43
Current status of the management of antimicrobial resistance in South Africa
Although there have been reports of the occurrence of antimicrobial-resistant genes in South African livestock and food items, the quantities of antimicrobials used are not yet monitored. Despite being banned a long time ago, some antimicrobial agents are still used in animal rearing (Table 1). Providing data on the consumption of antimicrobials is vital in the assessment and/or management of antimicrobial resistance, but pharmaceutical companies keep secret their data on the amount of antimicrobials sold in the South African market, not to mention the numerous varieties that are available to farmers over the counter.
South Africa is part of the Global Resistance Partnership (GARP) launched in February 2010. The intention of this Partnership is to address and analyse antimicrobial resistance issues in South Africa and partnering countries. Moreover, the South African National Veterinary Surveillance and Monitoring Programme for Resistance to Antimicrobial Drugs (SANVAD) was created alongside the South African Antibiotic Stewardship Programme (SAASP). Reports produced by these entities are still yet to be transcribed into fully operational policies and action plans. However, South Africa remains the most active African country as far as antimicrobial resistance surveillance is concerned. In this regard, the Global Action Plan on antimicrobial resistance initiated in partnership with the World Health Organization emphasises optimisation coupled with a strengthening of the knowledge and evidence base through surveillance and research on antimicrobial usage in human and animals. However, there still exists a paucity of consumption data worldwide, including in South Africa.44 Reports of the situational analysis of antibiotic use and resistance carried out in 2011 by South African stakeholders and the GARP through the Centre for Disease Dynamics Economics and Policy (CDDEP) project revealed many setbacks in the antimicrobial stewardship programmes; amongst which there was the unavailability of data from the intercontinental marketing services, causing a bias in the real picture of antimicrobial consumption in South Africa.
Regulation of antimicrobial usage in humans
Prescriptions in the public sector are guided by the Standard Treatment Guidelines which are based on the inclusion and the availability of the medicines on the Essential Medicines List.44 Prescriptions are unrestricted in the private sector with prescribers selecting whatever antimicrobials they feel are most appropriate. Provision is made through the Nursing Act 33 of 2005 under Section 56(6) in such a way that nurses can also prescribe to patients, especially in public HIV and TB healthcare centres. Act No. 53 of 1974 allows pharmacists to diagnose and prescribe antibiotics to patients if they suffer from common illnesses.45 A comparison of public and private sector data from the past 3 years from Intercontinental Marketing Services reveals an increase in the consumption of certain antibiotics (Table 2). This increase was a result of inappropriate use of antibiotics as first-line medicines and a lack of awareness of appropriate antibiotic prescribing or simply a willingness to use new drug formulations based on their availability on the market or based on the emergence of resistance to previous antibiotics.
Regulation of antimicrobial usage in animals
The Department of Agriculture, Forestry and Fisheries and the National Department of Health regulate the use of antibiotics by administering the Fertilizers, Farm Feeds, Agricultural Remedies and Stock Remedies Act (Act 36 of 1947) and the Medicines and Related Substances Control Act (Act 101 of 1965), respectively. The Stock Remedies Act 36 of 1947 was initiated to control the numerous parasites that were infesting South African livestock at the time. As time went by, certain antimicrobials, such as growth promoters, were allowed in order to help farmers in rural areas to access essential livestock medicines. The Medicines and Related Substances Control Act 101 of 1965 was initiated for prescription-only medicines and veterinary antibiotics are also controlled by this Act.
Measures to reduce the incidence and prevalence of vancomycin-resistant enterococci
Step 1: Enforcement of the legislation on drug distribution and usage
The dual registration process of medicines through the Agricultural Remedies and Stock Remedies Act (Act 36 of 1947) and the Medicines and Related Substances Control Act (Act 101 of 1965) presents some flaws and has raised concerns about the exacerbation of antimicrobial resistance emergence if there is no effective control. Stock remedies are distributed to veterinary wholesalers, distributors, farmers' cooperatives and feed mix companies by the manufacturer. Consequently, stock remedies are freely available and no record is kept of their use. Moreover, the South African situation deviates from the 1998 World Health Organization best practice guidelines in that: (1) the dual system of regulating veterinary products only partially addresses clear, transparent manufacturing requirements (whereas antibiotics listed under Act 101 of 1965 must be authorised with a Good Manufacturing Practice licence, stock remedies under Act 36 of 1947 are not); and (2) most authorised veterinary antibiotics are over-the-counter stock remedies and often administered by farmers. The World Health Organization recommends that only trained and licensed professionals decide when and how to use antibiotics.
Step 2: Prioritisation of the use of alternatives to antibiotics
Although vaccination campaigns are often costly, they could reduce the amounts of antimicrobials used in farming.46,47 Moreover, the use of probiotics, prebiotics and synbiotics in animal farming should be encouraged because these will improve the gut bacterial flora and therefore reduce disease occurrence that could necessitate antimicrobial usage.47,48 It has been shown that healthier gut microbial flora contribute to a better immune system with a better nutrient uptake and less colonisation by pathogens.49 Studies have demonstrated the usage of predatory bacteria to counter the pathogenic effects of the same strains or other pathogens after oral administration in chickens, cows and rabbits.50,51 Moreover, specific yeast strains with specific properties could be used as probiotics.52,53 Because of their specificity and their selectivity towards bacterial strains, bacteriophages represent another potential alternative to antimicrobial management of illnesses in animal farming even though they can be used in combination with some antimicrobials as there is no negative interaction between the two treatments.53-56 In addition, there is a tremendous amount of research on the potential use of antimicrobial peptides such as bacteriocins as an alternative to antibiotics.46,57 The South African research stakeholders and the pharmaceutical companies should therefore harness substantial research funding in these novel areas as they represent huge potential for the future.
Step 3: Implementation of a nationwide effective antimicrobial resistance surveillance system
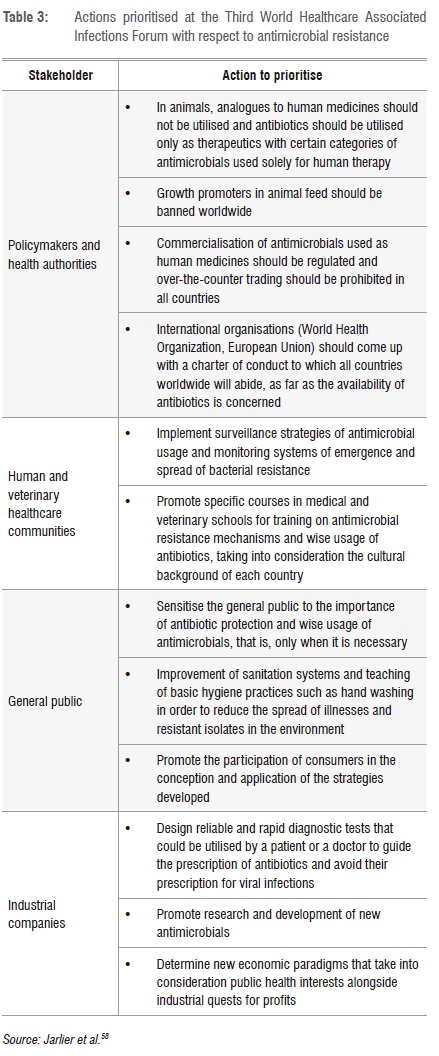
Large databases on antimicrobial resistance can be created and used for risk analyses and management in the different sectors of activity as well as in the different ecological niches that play a role in the dissemination and spread of resistant enterococci. Such surveillance systems have already been put in place in the European Union and in the USA to trace and control the patterns of antimicrobial distribution and that of the dissemination of antimicrobial-resistant bacteria. The database produced is playing a significant impact in the decision-making spheres in order to tackle antimicrobial resistance issues in the EU. This particular measure would be efficient if scientists and researchers were at the centre of the process and if international collaboration was encouraged because of the global aspect of antimicrobial resistance issues. In this regard, 70 experts from 33 countries gathered in 2011 at the Third World Healthcare Associated Infections Forum (WHAIF) that was dedicated to antibiotic resistance awareness and action.58 Actions that were agreed at this Forum are summarised in Table 3.
Last but not the least, the adoption of healthy lifestyles and the development of new drugs and vaccines should be encouraged as these constitute new perspectives as far as the therapeutic management of VRE infections is concerned.
Conclusion
Antimicrobials have been used in human and veterinary medicine ever since they were discovered. The intensive usage of antimicrobials in South Africa with their negative impact on food safety and human health has led to the emergence and the spread of VREs and other antimicrobial-resistant bacteria. In fact, epidemiological studies have demonstrated the link between antimicrobial usage and the detection of resistant isolates in the environment, making therapeutic alternatives for the treatment of infections caused by such organisms limited and their subsequent antimicrobial resistance genes available in the environment. As VREs are becoming a serious health threat in South Africa and worldwide, there is an urgent need to address this issue. Although the use of alternatives to antibiotics will effectively slow down the emergence process of resistant bacteria such as VREs, a synergistic approach involving the revision and the enforcement of laws and regulations on drug usage and distribution in South Africa, the encouragement of the invention of novel therapeutic molecules and the implementation of a critical nationwide antimicrobial resistance surveillance system will have significant impacts on the reduction of the prevalence or incidence of VREs as well as that of other antimicrobial-resistant bacteria. As awareness among the scientific community and the major stakeholders is increasing in South Africa, combined international action is required to tackle this issue once and for all.
Limitations
A limitation of this review was the lack of animal antimicrobial consumption data because of the unavailability or scarcity of data from wholesale suppliers and the Department of Agriculture, Forestry and Fisheries of South Africa.
Acknowledgements
We acknowledge the financial support of the North-West University towards the realisation of this study.
Authors' contributions
F.E.T.F. carried out the study and wrote the manuscript; A.K. and C.N.A. supervised the study and proofread the manuscript.
References
1. Butaye P, Devriese LA, Haesebrouck F. Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on gram positive bacteria. Clin Microbiol Rev. 2003;16(2):175-188. https://doi.org/10.1128/CMR.16.2.175-188.2003 [ Links ]
2. Marshall BM, Levy SB. Food animals and antimicrobials: Impacts on human health. Clin Microbiol Rev. 2011;24(4):718-733. https://doi.org/10.1128/CMR.00002-11 [ Links ]
3. Rosvoll TCS, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, et al. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol. 2010;58:254-268. https://doi.org/10.1111/j.1574-695X.2009.00633.x [ Links ]
4. Iweriebor BC, Gaqavu S, Obi CL, Nwodo UU, Okoh AI. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in Alice, Eastern Cape Province of South Africa. Int J Environ Res Publ Health. 2015;12:4231-4236. https://doi.org/10.3390/ijerph120404231 [ Links ]
5. Ateba CN, Maribeng MD. Detection of Enterococcus species in groundwater from some rural communities in the Mmabatho area, South Africa: A risk analysis. Afr J Microbiol Res. 2011;5(23):3930-3935. https://doi.org/10.5897/AJMR11.645 [ Links ]
6. Ateba CN, Lekoma KP, Kawadza DT. Detection of vanA and vanB genes in vancomycin-resistant enterococci (VRE) from groundwater using multiplex PCR analysis. J Water Health. 2013;11(4):684-691. https://doi.org/10.2166/wh.2013.037 [ Links ]
7. Taucer-Kapteijin M, Hoogenboezem W, Heiliegers L, Danny de Bolster H, Medema G. Screening municipal wastewater effluent and surface water used for drinking water production for the presence of ampicillin and vancomycin resistant enterococci. Int J Hyg Environ Health. 2016;16:1-7. [ Links ]
8. Kuzucu C, Cizmeci Z, Durmaz R, Durmaz E, Ozerol IH. The prevalence of faecal colonization of enterococci, the resistance of the isolates to ampicillin, vancomycin and high-level aminoglycosides and the clonal relationship among isolates. Microb Drug Resist Mech Epidemiol Dis. 2005;11(2):159-164. https://doi.org/10.1089/mdr.2005.11.159 [ Links ]
9. Sónia R, Ingrid C, Nuno S, Michel H, Hugo S, José-Luis CM, et al. Effect of vancomycin on the proteome of the multiresistant Enterococcus faecium SU18 strain. J Proteom. 2015;113:378-387. https://doi.org/10.1016/j.jprot.2014.10.012 [ Links ]
10. Jui-Chang T, Po-Ren H, Hsiao-Mann L, Hui-Jen C, Shen-Wu H, Lee-Jene T. Identification of clinically relevant Enterococcus species by direct sequencing of groES and spacer region. J Clin Microbiol. 2005;43:235-241. https://doi.org/10.1128/JCM.43.1.235-241.2005 [ Links ]
11. Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401-407. https://doi.org/10.1016/0966-842X(96)10063-9 [ Links ]
12. Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: Biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408-10415. https://doi.org/10.1021/bi00107a007 [ Links ]
13. Hughes D. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat Rev Genet. 2003;4:432-441. https://doi.org/10.1038/nrg1084 [ Links ]
14. Ateba CN, Mohapi MI. Isolation of vancomycin resistant enterococci isolated from leafy vegetables (lettuce) from North West Province. Life Sci. 2013;10(4):1163-1170. [ Links ]
15. Ruimy R, Brisabois A, Bernede C. Organic and conventional fruits and vegetables contain equivalent counts of Gram-negative bacteria expressing resistance to antibacterial agents. Environ Microbiol. 2010;12(3):608-615. https://doi.org/10.1111/j.1462-2920.2009.02100.x [ Links ]
16. Al-Kharousi ZS, Guizani N, Al-Sadi AM, Al-Bulushi IM, Shaharoona B. Hiding in fresh fruits and vegetables: Opportunistic pathogens may cross geographical barriers. Int J Microbiol. 2016; Art. #4292417, 14 pages. https://doi.org/10.1155/2016/4292417 [ Links ]
17. Matlou PD. Molecular analysis of vancomycin-resistant enterococci isolated from ground and surface water [MSc thesis]. Mafikeng: North-West University; 2016. [ Links ]
18. Molale LG. Characterization of enterococci isolates from water sources in the North West Province [PhD thesis]. Potchefstroom: North-West University; 2016. [ Links ]
19. Lochan H, Moodley C, Bamford C, Hendricks M, Davidson A, Eley B. Emergence of vancomycin-resistant Enterococcus at a tertiary paediatric hospital in South Africa. S Afr Med J. 2016;106(6):562-566. https://doi.org/10.7196/SAMJ.2016.v106i6.10858 [ Links ]
20. Mahabeer Y, Lowman W, Govind CN, Swe-Swe-Han K, Mlisana KP. First outbreak of vancomycin-resistant Enterococcus in a haematology unit in Durban, South Africa. S Afr J Infect Dis. 2016;31(1):20-24. https://doi.org/10.1080/23120053.2015.1118819 [ Links ]
21. Budavari SM, Saunders GL, Liebowitz LD, Khoosal M, Crewe-Brown HH. Emergence of vancomycin resistant enterococci in South Africa. S Afr Med J. 1997;87(11):1557. [ Links ]
22. Singh J, Esparza S, Patterson M, Vogel K, Patel B, Gornick W. Vancomycin-resistant Enteroococcus in paediatric oncology patients: Balancing infection prevention and family centered care. J Pediatr Hematol Oncol. 2013;35(3):227-231. https://doi.org/10.1097/MPH.0b013e318257a6ca [ Links ]
23. Ergani-Ozcan A, Naas T, Baysan BO, Ogunc D, Inan D, Colak D, et al. Nosocomial outbreak of vancomycin-resistant Enterococcus faecium in a paediatric unit at a Turkish university hospital. J Antimicrob Chemother. 2008;61(5):1033-1039. https://doi.org/10.1093/jac/dkn066 [ Links ]
24. Isogai N, Urushibara N, Kawaguchiya M, Ghosh S, Suzaki K, Watanabe N, et al. Characterization of Enterococcus faecium with macrolide resistance and reduced susceptibility to quinupristin/dalfopristin in a Japanese hospital: Detection of extensive diversity in erm(B)-regulator regions. Microb Drug Resist. 2013;19:298-307. https://doi.org/10.1089/mdr.2012.0176 [ Links ]
25. Moges F, Endris M, Belyhun Y, Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, northwest Ethiopia. BMC Res Notes. 2014;7:1-6. https://doi.org/10.1186/1756-0500-7-215 [ Links ]
26. Ngu TG. Genotypic and phenotypic characterization of enterococci from cow dung and environmental water sources in three selected dairy farms in Amathole District [MSc thesis]. Alice: University of Fort Hare; 2016. [ Links ]
27. Moyane JN, Jideani AIO, Aiyegoro OA. Antibiotic usage in food producing animals in South Africa and impact on human: antibiotic resistance. Afr J Microbiol Res. 2013;7(24):2990-2997. https://doi.org/10.5897/AJMR2013.5631 [ Links ]
28. Iweriebor BC, Obi CL, Okoh AI. Virulence and antimicrobial resistance factors of Enterococcus spp. isolated from faecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol. 2015;15, Art. #136, 11 pages. https://doi.org/10.1186/s12866-015-0468-7 [ Links ]
29. Jackson CR, Fedorka-Cray PJ, Barrett JB, Ladely SR. High-level aminoglycoside resistant enterococci isolated from swine. Epidemiol Infect. 2005;133:367-371. https://doi.org/10.1017/S0950268804003395 [ Links ]
30. Aarestrup FM, Kruse H, Tast E, Hammerum AM, Jensen LB. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb Drug Resist. 2000;6:63-70. https://doi.org/10.1089/mdr.2000.6.63https://doi.org/10.1089/mdr.2000.6.71 [ Links ]
31. Boxall AB, Blackwell P, Cavallo R, Kay P, Tolls J. The sorption and transport of a sulphonamide antibiotic in soil systems. Toxicol Lett. 2002;131:19-28.https://doi.org/10.1016/S0378-4274(02)00063-2 [ Links ]
32. Ding GC, Radl V, Schloter-Hai B, Jechalke S, Heuer H, Smalla K, et al. Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS One. 2014;9(3), e92958, 10 pages. https://doi.org/10.1371/journal.pone.0092958 [ Links ]
33. Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612-616. https://doi.org/10.1038/nature13377 [ Links ]
34. Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. MBio. 2016;7(2), e02227-15, 7 pages. https://doi.org/10.1128/mBio.02227-15 [ Links ]
35. Drissner D, Zürcher U. Microbial safety of fresh fruits and vegetables. In: Motarjemi Y, editor. Encyclopedia of food safety. Vol 3. Oxford: Elsevier; 2014. https://doi.org/10.1016/B978-0-12-378612-8.00280-8 [ Links ]
36. Soonthornchaikul N, Garelick H. Antimicrobial resistance of Campylobacter species isolated from edible bivalve molluscs purchased from Bangkok markets, Thailand. Foodborne Pathog Dis. 2009;6(8):947-951. https://doi.org/10.1089/fpd.2008.0236 [ Links ]
37. Petersen A, Andersen JS, Kaewmack T, Somsiri T, Dalsgaard A. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl Environ Microbiol. 2002;68(12):6036-6042. https://doi.org/10.1128/AEM.68.12.6036-6042.2002 [ Links ]
38. Shah SQA, Colquhoun DJ, Nikuli HL, Sørum H. Prevalence of antibiotic resistance genes in the bacterial flora of integrated fish farming environments of Pakistan and Tanzania. Environ Sci Technol. 2012;46(16):8672-8679. https://doi.org/10.1021/es3018607 [ Links ]
39. Rossi F, Rizzoti L, Felis GE, Torriani S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 2014;42:232-243. https://doi.org/10.1016/j.fm.2014.04.004 [ Links ]
40. Toomey N, Monaghan A, Fanning S, Bolton DJ. Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog Dis. 2009;6(8):925-933. https://doi.org/10.1089/fpd.2009.0278 [ Links ]
41. Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1991;35(1):185-187. https://doi.org/10.1128/AAC.35.1.185 [ Links ]
42. Rizzotti L, La Gioia F, Dellaglio F, Torriani S. Molecular diversity and transferability of the tetracycline resistance gene tet(M), carried on Tn916-1545 family transposons, in enterococci from a total food chain. Antimicrob Agents Chemother. 2009;96(1):43-52. https://doi.org/10.1007/s10482-009-9334-7 [ Links ]
43. Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49-61. https://doi.org/10.2147/IDR.S55778 [ Links ]
44. Perumal-Pillay V, Suleman F. Selection of essential medicines for South Africa - an analysis of in-depth interviews with national essential medicines list committee members. BMC Health Serv Res. 2017;17, Art.#17, 17 pages. https://doi.org/10.1186/s12913-016-1946-9 [ Links ]
45. Schellack N, Deon B, Brink A, Duse A, Faure K, Goff D, et al. A situational analysis of current antimicrobial governance, regulation, and utilization in South Africa. Int J Infect Dis. 2017;64:100-106. https://doi.org/10.1016/j.ijid.2017.09.002 [ Links ]
46. Allen HK, Trachsel J, Looft T, Casey TA. Finding alternatives to antibiotics. Ann NY Acad Sci. 2014;1323(1):91-100. https://doi.org/10.1111/nyas.12468 [ Links ]
47. Callaway TR, Edrington TS, Anderson RC. Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim Health Res Rev. 2008;9(2):217-225. https://doi.org/10.1017/S1466252308001540 [ Links ]
48. Gaggìa F, Mattarelli P, Biavati B. Probiotics and prebiotics in animal feeding for safe food production. Int J Food Microbiol. 2010;141(1):15-28. https://doi.org/10.1016/j.ijfoodmicro.2010.02.031 [ Links ]
49. Seal BS, Lillehoj HS, Donovan DM, Gay CG. Alternatives to antibiotics: A symposium on the challenges and solutions for animal production. Anim Health Res Rev. 2013;14(1):78-87. https://doi.org/10.1017/S1466252313000030 [ Links ]
50. Atterbury RJ, Hobley L, Till R. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol. 2011;77(16):5794-5803. https://doi.org/10.1128/AEM.00426-11 [ Links ]
51. Dwidar M, Monnappa AK, Mitchell RJ. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012;45(2):71-78. https://doi.org/10.5483/BMBRep.2012.45.2.71 [ Links ]
52. Biliouris K, Babson D, Schmidt-Dannert C, Kaznessis YN. Stochastic simulations of a synthetic bacteria-yeast ecosystem. BMC Syst Biol. 2012;6(1), Art. #58, 13 pages. http://dx.doi.org/10.1186/1752-0509-6-58 [ Links ]
53. Kenny M, Smidt H, Mengheri E, Miller B. Probiotics - do they have a role in the pig industry? Animal. 2011;5(3):462-470. https://doi.org/10.1017/S175173111000193X [ Links ]
54. Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013;8(6):769-783. https://doi.org/10.2217/fmb.13.47 [ Links ]
55. Goodridge LD, Bisha B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage. 2011;1(3):130-137. https://doi.org/10.4161/bact.1.3.17629 [ Links ]
56. Balogh B, Jones JB, Iriarte FB, Momol MT. Phage therapy for plant disease control. Curr Pharm Biotechnol. 2010;11(1):48-57. https://doi.org/10.2174/138920110790725302 [ Links ]
57. Snyder AB, Worobo RW. Chemical and genetic characterization of bacteriocins: Antimicrobial peptides for food safety. J Sci Food Agric. 2014;94(1):28-44. https://doi.org/10.1002/jsfa.6293 [ Links ]
58. Jarlier V, Carlet J, McGowan J, participants of the 3rd World Healthcare-Associated Infections Forum. Priority actions to fight antibiotic resistance: Results of an international meeting. Antimicrob Resist Infect Control. 2012;1(1), Art. #17, 3 pages. https://doi.org/10.1186/2047-2994-1-17 [ Links ]
 Correspondence:
Correspondence:
Frank Tatsing Foka
Email: frankeric2023@yahoo.fr
Received: 05 Oct. 2017
Revised: 06 Dec. 2017
Accepted: 07 May 2018
Published: 11 Sep. 2018
Funding: North-West University














