Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.114 no.7-8 Pretoria jul./ago. 2018
http://dx.doi.org/10.17159/sajs.2018/20170018
RESEARCH ARTICLE
Community composition and functions of endophytic bacteria of Bt maize
Asnath R. MashianeI, II; Rasheed A. AdelekeI, II; Cornelius C. BezuidenhoutII; George J. ChirimaIII
IMicrobiology and Environmental Biotechnology Research Group, Agricultural Research Council, Pretoria, South Africa
IIUnit for Environmental Sciences and Management, North-West University, Potchefstroom, South Africa
IIICenter for African Ecology, University of the Witwatersrand, Johannesburg, South Africa
ABSTRACT
We investigated the potential effects of genetic modification of Bt maize on the community composition and functions of bacterial endophytes associated with transgenic maize (Bt MON 810) in comparison with its isogenic parental line at two developmental stages. Bacterial isolates were obtained from transgenic (Bt) and non-transgenic (non-Bt) maize at 50- and 90-day-old developmental stages. Isolated bacterial endophytes were screened for their capabilities in phosphate solubilisation, nitrogen fixation, production of antifungal metabolites and production of indole acetic acid. After molecular identification, 60 isolates were obtained and clustered into 19 and 18 operational taxonomic units from 50- and 90-day-old maize, respectively. The isolates belonged to the genera Bacillus, Pantoea, Serratia, Yersinia, Enterobacter, Pseudomonas, Acinetobacter and Stenotrophomonas. Functional attributes and diversity of the isolated endophytes at both developmental stages were not significantly different for both maize varieties. However, functional attributes were significantly affected by plant growth stage. Isolates from younger plants were more efficient producers of indole acetic acid, but exhibited little or no capabilities for nitrogen fixation, phosphate solubilisation and antifungal activity in both maize genotypes. Based on these outcomes, Bt modification in maize does not seem to affect the community composition or functional attributes of bacterial endophytes.
SIGNIFICANCE:
•Bt modification in maize does not affect the ecological guild or functional attributes of cultivable bacterial endophytes.
Keywords: endophytes; nitrogen fixation; phosphate solubilisation; antifungal activity; transgenic (Bt) maize
Introduction
Maize is one of the most important crops in the world, and is consumed as a staple food as well as animal feed in both developed and developing countries.1 Globally, maize production is threatened by a number of factors, especially the outbreak of pests such as stalk borer insects.1 The tissue damage caused by the stem borers allows pathogenic microbes to colonise open areas leading to leaf and cob rots as well as mycotoxin accumulation.1 The introduction of genetically modified Bt maize, which contains a gene from the soil bacterium Bacillus thuringiensis (Bt), has given rise to the production of highly resistant varieties that are toxic to major insect pests of the orders Lepidoptera and Coleoptera.2,3 On the other hand, the overall impact of such modification in maize cannot be predicted.4 For example, such genetic modification could adversely affect non-target organisms, especially the composition and functions of the natural microbial community of maize plants, which includes endophytic, ectophytic and rhizospheric microbes.5,6
Endophytes are bacteria, archaea or fungi that live inter- or extracellularly without causing any symptoms of disease to the host plant.7 They are present in virtually all plants studied to date.8-10 Their diversity varies from plant to plant as a result of many factors that may include, among others, plant species, genotype, tissue, growth stage and differences in colonisation pathway.6,11 Bacterial endophytes have the capacity to promote plant growth because of their participation in nutrient cycling - phosphorus solubilisation and nitrogen fixation, hormone production (indole acetic acid) as well as suppression of pathogens (biocontrol agents).12-16 Gaining a more detailed understanding of the microbial community in genetically modified maize is imperative for evaluating the resilience of varieties as well as the potential implication of the modification on ecosystem functioning. Few studies have delved into this important aspect of endophyte biology.3,17-19 In this study, we hypothesised that genetic modification of maize plants (such as Bt maize) might influence the diversity and functional attributes of endophytic bacteria associated with the plant. This led us to investigate the potential impacts of genetic modification of maize on the community composition and functions of cultivable bacterial endophytes from different plant parts that included leaves, stems, cobs and tassels/husks at two different plant growth stages. Such an approach will improve our knowledge in this field, particularly that relating to: (1) the relationship between maize phyllosphere and community composition of bacterial endophytes and (2) potential impacts of genetic modification of Bt maize on the community composition and functional roles of the bacteria endophytes at different plant growth stages.
Methods and materials
Sample collection
Samples were collected at the Agricultural Research Council - Grain Crop Institute in Potchefstroom in the North West Province of South Africa (26°43'39.2'S, 27°04'48.8'E). The study was conducted using a single variety of transgenic Bt maize (MON 810) and its isogenic parental line (non-Bt), which served as a reference or control. The Bt and non-Bt maize fields were cultivated using disc ploughing. The maize was hand planted in 1.2-m rows. The soil type for both plant genotypes was the same as the plots were next to each other. Plants were irrigated weekly as needed. Field temperatures varied between 24 °C and 32 °C. Plants were sampled 'destructively' at two developmental stages: pre-flowering (50 days after emergence) and post-flowering (90 days after emergence). A total of 20 maize plants (10 Bt and 10 non-Bt) were sampled and analysed at each developmental stage. The plant parts were severed with sterile scissors, placed in a plastic bag and transported to the laboratory. Collected explants of healthy leaves, stem, tassels and seeds were stored separately for analysis.
Isolation of endophytes
Stored samples of healthy leaves, stem, tassels and seeds were cut into pieces of approximately 20 mm2 segments (explants). The explants were surface sterilised using a three-step approach that involved immersion in 70% (v/v) ethanol for 60 s, followed by rinsing with distilled water and subsequent sterilisation in 3% (v/v) sodium hypochlorite for 60 s and finally in 70% (v/v) ethanol for 30 s. Samples were further washed in sterile distilled water three times, for 60 s each.20 Nutrient agar (Merck (Pty) Ltd, Johannesburg, South Africa), tryptone soy agar (Merck), and nutrient broth media were used for the isolation of bacteria from the explants. The process involved the inoculation of explants obtained from different parts of the maize plants on the three different media. The explants were inoculated at the centre of the plates containing these three media. All plates were incubated at 27 °C in duplicate for 24 h. Sub-culturing was done until pure isolates were obtained. Pure isolates from the same plant parts but different replicates were grouped together in subsequent analyses.
Colony PCR and sequencing
An aliquot of pure single colony culture was transferred to a 1.5-mL microcentrifuge tube containing 200 µL sterile milliQ water and homogenised using a vortex (Labnet International, Edison, NJ, USA). DNA amplification of the isolates was done directly using the suspended cells in a colony polymerase chain reaction (PCR).21,22 The PCR targeted the partial 16S rDNA gene of the bacterial isolates. The amplification was done using primer sets 341F (5' CCTACGGGAGGCACCAG3') and 907R (5' CCGTCAATTCCTTTGATTT3').23 Primers were synthesised by Inqaba Biotech (South Africa). The 20 μL reaction mixture included 2X PCR master mix (0.05 U/μL Phusion Flash II DNA polymerase, 4 mM MgCl2 and 0.4 mM dNTPs (Thermo scientific, USA), specific primers (100 pmole/μL) and distilled water). Reagents were mixed by brief centrifugation at 6500 rcf (Cencom I, Barcelona, Spain). The PCR programme involved an initial denaturation step of 98 °C for 60 s, 35 cycles of 98 °C for 30 s, 50 °C for 30 s and 72 °C for 60 s, with a final extension step of 72 °C for 5 min. The integrity of PCR amplicons was verified with the aid of gel electrophoresis on a 1% agarose gel using a 100-bp molecular marker (Biolabs). Amplicons (≈600 bp) were further purified and sequenced bi-directionally with the same set of primers. Forward and reverse sequences were inspected, edited and assembled using Bio-Edit.24 Sequence data obtained were submitted to Genbank and given accession numbers KT120070-KT120099 and KT459755-KT459782.
Taxonomic assignment and phylogenetic reconstruction
All the 16S rDNA gene sequences were assigned to bacterial taxa using SeqMatch on the Ribosomal Database Project website (http://rdp.cme.msu.edu/index.jsp).25 This assignment was followed by multiple sequence alignments and clustering into operational taxonomic units (OTUs) using the mothur software.26 The assignment was based on a 99% similarity level between OTUs. Matched sequences, one for each OTU, were later obtained from the National Center for Biotechnology Information (NCBI)'s Genbank using the accession numbers. These sequences alongside the OTU representatives were used to construct a library. All sequences were aligned using the multiple sequence alignment software MAFFT version 7.27 Mega6 software was used to generate a neighbour-joining phylogenetic tree consisting of representative OTUs and their close relatives (matched sequences).28
Nitrogen fixation and indole acetic acid assay
Isolates were streaked on a Burk's nitrogen-free culture medium. The medium comprised 10 g glucose, 0.52 g K2HPO4, 0.41 g KH2PO4, 0.05 g Na2SO4, 0.2 g CaCl2, 0.1 g MgSO4·7H2O, 0.005 g FeSO4·7H2O, 0.0025 g Na2MoO4·2H2O, 15 g agar per litre.29 Inoculated plates were incubated at 28 °C for 7 days to allow growth. Bacterial isolates showing visible growth on the plates were considered to be positive nitrogen fixers.30
For indole acetic acid (IAA) production, the cultures (in triplicate) were grown on tryptophan broth and incubated at 30 °C for 48 h on a shaker incubator at 180 rcf.31 Bacterial cells were separated from the supernatant by centrifugation at 10 000 rcf for 10 min in a centrifuge (Biocen 22 R, Orto Alresa, Madrid, Spain). IAA was measured by mixing 1 mL broth with 2 mL Salkowsky's reagents (2% of 0.5 M FeCl3 + 35% HClO4), and the resulting suspension was incubated for 20 min at room temperature. Absorbance was measured at 530 nm using a spectrophotometer (V-1100D, Xinke Instruments Co., Ltd, Sichuan, China). The final amount of IAA was calculated using the standard of pure IAA (Sigma-Aldrich Ltd, Johannesburg, South Africa) prepared separately. IAA production was indicated by development of a pink colouration.
Phosphate solubilisation
The phosphate solubilisation ability of the bacterial isolates was assessed by plate assay using the Indian National Botanical Research Institute's phosphate growth medium.32,33 The medium contained (g/L): MgSO4.7H2O-0.25; (NH4)2SO4-0.10; MgCl2.6H2O-5.00; KCl-0.20; Ca3(PO4)2-2.5; glucose-10 and agar-20. A clear halo around the bacterial colony was considered positive for phosphate solubilisation. Each of the isolates was inoculated in triplicate and grown at 30 °C for 8 days.
The negative control was without the inoculum.
Solubilisation efficiency = (diameter of halo)/ (diameter of colony) X 100
Antifungal activity assay
The maize pathogen used for the antifungal activity assay in this study was Fusarium verticillioides 10025, obtained from the Agricultural Research Council - Plant Protection Research Institute. The test was performed using potato dextrose agar (Merck (Pty) Ltd, South Africa) medium that had a 5-mm disc of fungi mycelia (F. verticillioides 10025) placed at the centre of the plate, with a single streak of bacterial culture 3 mm away from the fungal disc. The plates were grown for 5 days at 25 °C and the inhibition activity was evaluated by comparing the radius of the fungal growth treated with bacteria against the control. The inhibition estimation was calculated by:
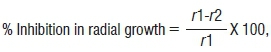
where r1 is the radial mycelia growth in the control and r2 is the radial mycelia growth in the treatment. The antifungal activity of each isolate was tested in triplicate.
Statistical analyses
Assessments of potential impacts of genetic modification of the BT maize on the type, composition and functions of endophytes prevalent at each growth stage were determined by multivariate analysis.34-36 For this analysis, data for all plant parts were pooled per growth stage.
Multivariate analysis combines different measurements from the same sample and can recognise correlations and interactions between factors; it is therefore a good tool to understand external influences on species composition as well influences on functions simultaneously.34 For this reason, many studies aiming to quantify the effects of transgenic plants on microbial-associated communities have used this instrument.37
Community composition as well as capabilities of the endophytes for solubilisation of phosphate, fixation of atmospheric nitrogen, production of antifungal metabolites, and production of IAA were compared between Bt and non-Bt maize genotypes for the pre- and post-flowering stages. Principal component analysis (PCA) was used to determine how the isolated endophytes could be related directly to each of the identified functional attributes. This approach not only allowed the detection of the variation in community composition but also showed their functional capabilities at both the pre-flowering stage (50 days) and post-flowering stage (90 days). The PCA was applied in MS Excel version 2013. Data were arranged such that the four variables - nitrogen fixation, % inhibition, % phosphate efficiency and IAA production - were mapped as arrows. In the resulting bi-plot, endophyte types were represented by points, circles, triangles etc., while function variables were represented by arrows. Bi-plots represented an endophyte's position along an arrow representing a capability to perform that particular function, because arrows representing each function were plotted in the direction of their maximum change. Thus, long arrows indicated high capability for that function. Numbers (diversity) of isolated endophytes from Bt versus non-Bt maize did not differ if points occurred close together or in the same positions along the arrows. Endophyte species occurring in positions close to or beyond the tip of the specific arrow were strongly and positively correlated with that functional capability. A perpendicular from the arrow to an endophyte point indicated the position of that species in relation to values of that function. Those species at the opposite end of the arrow were less strongly affected.38 Thus, in addition to presenting the variation in community composition, the bi-plot also accounted for variation in ability to solubilise phosphate, fix atmospheric nitrogen, and produce antifungal metabolites.36,39 In this way, the PCA approach allowed a quick appraisal of how community composition and functions vary between the pre- and post-flowering stages and between the Bt and non-Bt maize varieties.39
Statistical tests were run on SPSS version 19.0 comparing these capabilities during the two periods - pre-flowering and post-flowering. For the capacity to fix nitrogen during pre-flowering versus post-flowering, a score of 1 was applied if nitrogen fixation was present and 0 if absent, and the analysis was conducted using chi-square tests. For solubilisation of phosphate, production of IAA, and antifungal activity at 50 days, independent t-tests were applied (Supplementary table 1). Because endophyte activity at 90 days was not independent of endophyte activity at 50 days, paired t-tests were used to compare these activities between the two treatments at 90 days (Supplementary table 2). To broadly assess functional efficiencies of the species between plant growth stages, a one-way repeated measures analysis of variance comparing the capabilities for specific function at each growth stage was applied.
Results
Bacterial isolates
The amplified DNA products were 600 bp in length. The sequences were clustered into OTUs at 99% similarity. The homology sequence and phylogenetic analyses of the 16S rDNA of the bacterial endophytes associated with stems, leaves, tassels and seeds of Bt and non-Bt maize indicated that they belong to eight genera: Bacillus, Pantoea, Stenotrophomonas, Yersinia, Serratia, Pseudomonas, Enterobacter and Acinetobacter (Figures 1 to 4; Table 1).
Table 1 indicates the number of isolates obtained from both Bt (transgenic) and non-Bt maize plants with their accession numbers and OTU representatives. The diversity of cultivable bacterial endophytes (Figures 3 and 4) in the Bt variety was not significantly different from that of the non-Bt maize at 50 days. The prevalence of species at 50 days in Bt maize was 48.3%, whilst in non-Bt maize was 51.7%. The relative prevalence of species in Bt maize increased as a function of growth of the plants from 48.3% to 66% at 90 days (Figures 1 and 2). The diversity of endophytes obtained in the present study were of the phyla Firmicutes (28%) and Proteobacteria (72%). From the endophytic bacterial species obtained from both maize genotypes, Bacillus was the most recurrent from the leaves (17%), stem (12%) and seeds (8%) while Pantoea was isolated more from the leaves (12%). The genus Bacillus was found in all the plant parts investigated while Pantoea and Enterobacter were found in the stems and leaves. Stenotrophomonas and Serratia were identified from the seeds and stems. Lastly, Yersinia was identified in the leaves and tassels while Acinetobacter was only obtained from the stems (Table 1).
Bacterial isolates and functions
Bacterial community composition between Bt and non-Bt maize was not different, suggesting that Bt modification may not have a negative impact on the bacterial endophyte populations. Specifically, there was no significant difference in terms of endophyte community composition between Bt and non-Bt for both plant growth stages, pre-flowering and post-flowering (Figure 5). Lower capability for nitrogen fixation was strongly associated with the pre-flowering stage (i.e. young plants) for both Bt and non-Bt maize (Figure 6). However, all isolates from the post-flowering stage were able to fix nitrogen in both Bt and non-Bt maize (Figure 6). Antifungal activity and phosphate solubilisation capabilities of the isolates were positively associated with the post-flowering stage and weakly associated with the pre-flowering stage.
For the pre-flowering plants, on both non-Bt and Bt maize, the highest IAA production was particularly by BT4_50S (Pantoea species) and NBT5_50S (Acinetobacter species). However, there were three species with exceptions to the growth stage trend as indicated in Figure 5a. These endophytes exhibited high antifungal activity and phosphate efficiency.
Screening for potential atmospheric nitrogen fixers
There were no significant differences in nitrogen fixation between Bt and non-Bt maize at 50 days (p=0.779). The number of nitrogen fixers increased in both Bt and non-Bt varieties from 50 days to 90 days; the increase was significant in the Bt variety (p=0.003).
Indole acetic acid assay
Figure 7 shows the activity of IAA production at both 50 days and 90 days for Bt and non-Bt maize plants. There were no significant differences in IAA production between Bt and non-Bt maize at 50 days (p=0.555). However, there was a significant difference in IAA production between 50 and 90 days' developmental growth stages for both Bt and non-Bt maize varieties, with isolates obtained from younger plants exhibiting an elevated capacity for production of IAA (p=0.008).
Phosphate solubilisation
The distribution of phosphate solubilisation is presented in Figure 8. Phosphate solubilisation was not significantly different for Bt and non-Bt maize varieties compared laterally at both 50- and 90-day periods (p= 0.582) (compared independently). However, when compared across growth stages, isolates obtained from older plants (post-flowering) were significantly more efficient in phosphate solubilisation (p=0.0027).
Antifungal activity
Figure 9 illustrates the bacterial endophytes with potential inhibition against the test fungal phytopathogen (F. verticillioides). There was no significant difference between Bt and non-Bt maize in terms of antifungal activity at 50 (p=0.627) and 90 days (p=0.652) when compared separately. However, when plant age was considered for both categories of Bt and non-Bt maize, antifungal activity was highest in older plants (i.e. post-flowering) (p=0.001).
Discussion
Transgenic Bacillus thuringiensis (Bt) maize is the most widely grown crop in the world.40 The transgenic plants have functional genes inserted into their genome which are expressed in all the tissues and stages during plant growth. When such genetic modification exists in the plant tissues, non-target organisms such as endophytes, epiphytes and rhizospheric microbes could be at risk. Unfortunately, plants greatly depend on these microbes for health and growth as they play a crucial role in nutrient mineralisation, biological control, hormone production and resistance to stress.5 Possible effects of the genetic modification (Bt maize) on the community composition and functional attributes of endophytes during two developmental plant growth stages in comparison to the non-Bt maize isogenic parental line were evaluated. Remarkably, there was no significant difference in the composition of the culturable bacterial community of the two maize genotypes. This result could be related to maize plants having the same physiological characteristics, thus hosting the same endophytic group. Similarly, another study done by Saxena and Stotzky5 did not find any difference in bacterial communities between transgenic and non-transgenic maize. However, shifts in microbial density of Bt maize were observed during the reproductive stage (90 days old), when compared to non-Bt maize, which could be related to plant age and soil type.11,41
The phylogenetic pattern of culturable endophytes obtained in the present study revealed that they belong to the phyla Firmicutes and Proteobacteria. This finding is in agreement with previous studies linking these bacterial phyla to maize.18,42 It has also been reported that, generally, Gamma- and Alphaproteobacteria are the dominant bacterial community inhabiting the phyllosphere, although the Firmicutes and Betaproteobacteria can also be present in large numbers. In this study, the dominant bacterial inhabitants were the Gammaproteobacteria and Firmicutes (Figures 3 and 4). The high density of endophytes on leaves compared to other plant parts might be because leaves have a large surface area and natural openings (stomata), allowing them to be the preferred point of tissue entry, as observed by Kumar and Hyde43. Furthermore, it has been shown that different species are commonly found on multiple plant tissues and there are some with a preference for the leaves.15,19,44 This shows that there is a correlation between endophytes and plant parts harbouring them. In the present study, Bt and non-Bt maize cultivars harboured unrelated genera that are commonly found as maize endophytes such as Pantoea, Bacillus, Enterobacter, Serratia, Yersinia, Stenotrophomonas, Pseudomonas and Acinetobacter (Table 1).18,42 The results revealed the predominance of Bacillus and Pantoea, as well as the overall importance of all the isolated endophytes in plant growth processes. PCA clearly confirms that there is no differentiation between Bt and non-Bt maize. Endophyte functional diversity, which represents the capacity of microorganisms to perform different biological and ecological processes, is an important indicator of system disturbance and development. The PCA showed that the microbial community in the Bt and non-Bt maize at 50 days and at 90 days was not distinct.
Plant growth is sustained by the soil fertility state and a fertile soil is defined by the presence of important nutrients such as nitrogen, phosphorus and potassium.45 Although nitrogen fixation has always been associated with legumes, it has been shown that non-leguminous plants such as maize also benefit from nitrogen fixers.46 The most important constraint in maize production is low soil nitrogen, which contributes to a loss in production of about 30%.47 Hence the presence of nitrogen fixers in the soil serves as a potential source of replacement for such loss. Furthermore, most of the potential nitrogen fixers in the rhizosphere have additional beneficial roles. For instance, in addition to their nitrogen-fixing capabilities, nitrogen fixers can also participate in root expansion, bioremediation and nutrient cycling.46,48,49 In the present study, the number of isolates with nitrogen-fixing capabilities for both maize varieties (Bt and non-Bt) increased significantly between the two developmental stages (Figure 5a). The PCA showed that isolates with nitrogen-fixing capabilities for both maize varieties (Bt and non-Bt) clustered together (Figure 5b), with more isolates from the 90-day-old developmental stage. This probably occurs because more nitrogen is needed during maximum plant growth for reproduction purposes and as the plants continue to grow, available nitrogen becomes depleted.47 The genera which had significant influence in fixing atmospheric nitrogen were Bacillus and Pantoea.
IAA production by isolates was significantly higher at 50 days than at 90 days in both maize varieties. As shown in Figure 5a, only 50-day isolates of both maize varieties grouped together. The major reason for clustering or high IAA at 50 days appears to be associated with plant age because it is a growth hormone needed by the plants at an early stage for root and stem growth regulation.48 Similarly, the same pattern was observed with regard to phosphate solubilisation and antifungal activity, with an increase in both functional traits associated with isolates at the 90-day developmental stage.
The results obtained in this study are important for both scientists and farmers, especially as they relate to different biotechnological applications. Globally, the ultimate goal of farmers is to cultivate maize of desirable traits. Important scientific information that indicates that Bt maize does not have negative impacts on the ecosystem will be welcomed. This information may in turn translate to improvements in acceptability and marketability of the maize being produced by the farmers, as well as the ability to invest in environmentally friendly fertilisation approaches (e.g. biofertiliser) which rely on microbial activities.
A limitation of the study is the number of genotypes used as well as the sole use of a culture-based method. However, the culture-based approach was important in this study because it allowed us to assess the functional traits of the isolated bacterial endophytes (Figure 5b). In comparison, Mashiane et al.'s49 metagenomics analytical approach provided more information about the abundance and diversity of the bacterial endophytes.
In conclusion, we have demonstrated that maize phyllosphere harbours different types of bacterial endophytes but their composition is not affected by the Bt genetic modification of the maize plant. Similarly, functional roles of the bacterial endophytes are not affected by the genetic modification. However, there was a significant increase in endophyte density from the 50-day to the 90-day developmental stage, suggesting that developmental stages of both Bt and non-Bt maize could drive the composition of the endophytic bacterial community. The beneficial characteristics of the endophytic bacteria in this study are important in agriculture. Thus, further biotechnological investigation needs to be conducted under field conditions to confirm the efficiency of these bacterial isolates in nutrient cycling and plant protection.
Acknowledgements
This study was funded by the National Research Foundation (NRF) through the IRG - Egypt/South Africa Research Cooperation Programme (grant no.108673) awarded to R.A.A. We acknowledge the Agricultural Research Council's sponsorship of A.R.M. through the professional development programme of the Agricultural Research Council. We sincerely thank Mr Owen Rhode and members of the Microbiology and Environmental Biotechnology Research Group, ARC-ISCW for their technical assistance.
Authors' contributions
A.R.M.: Methodology, data collection, data analysis, sample analysis, validation, data curation, writing the initial draft. R.A.A.: Conceptualisation, methodology, data collection, data analysis, sample analysis, validation, data curation, writing the initial draft and revisions, student supervision, project leadership, project management, funding acquisition. C.C.B.: Methodology, sample analysis, validation, writing the initial draft and revisions. G.J.C.: Data analysis, validation.
References
1. Keetch DP, Webster JW, Ngqaka A, Akanbi R, Mahlangu P. Bt maize for small scale farmers: A case study. Afr J Biotechnol. 2005;4(3):1505-1509. http://doi:10.4314/ajfand.v4i13.71805 [ Links ]
2. Ostry V, Ovesna J, Skarkova J, Pouchova V, Ruprich J. A review on comparative data concerning Fusarium mycotoxins in Bt maize and non-Bt isogenic maize. Mycotoxin Res. 2010;26(3):141-145. http://dx.doi.org/10.1007/s12550-010-0056-5 [ Links ]
3. Prischl M, Hackl E, Pastar M, Pfeiffer S, Sessitsch A. Genetically modified Bt maize lines containing cry3Bb1, cry1A105 or cry1Ab2 do not affect the structure and functioning of root-associated endophyte communities. Appl Soil Ecol. 2012;54:39-48. http://doi.org/10.1016/j.apsoil.2011.12.005 [ Links ]
4. Sticher L, Mauch-Mani B, Métraux JP. Systemic acquired resistance. Annu Rev Phytopathol. 1997;35(1):235-270. http://doi.org/10.1146/annurev.phyto.35.1.235 [ Links ]
5. Saxena D, Stotzky G. Insecticidal toxin from Bacillus thuringiensis is released from roots of transgenic Bt corn in vitro and in situ. FEMS Microbiol Ecol. 2000;33(1):35-39. https://doi.org/10.1111/j.1574-6941.2000.tb00724.x [ Links ]
6. Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67(2):257-268. https://doi.org/10.1021/np030397v [ Links ]
7. Das A, Varma A. Symbiosis: The art of living. Symbiotic fungi. In: Varma A, Kharkwal AC, editors. Symbiotic fungi principles and practice. Berlin: Springer; 2009. p. 1-28. https://doi.org/10.1007/978-3-540-95894-9_1 [ Links ]
8. Nair DN, Padmavathy S. Impact of endophytic microorganisms on plants, environment and humans. Sci World J. 2014;2014:1-11. http://dx.doi.org/10.1155/2014/250693 [ Links ]
9. Ryan PR, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: Recent developments and applications. FEMS Microbiol Lett. 2008;278(1):1-9. https://doi.org/10.1111/j.1574-6968.2007.00918.x [ Links ]
10. Lee S, Flores-Encarnación M, Contreras-Zentella M, Garcia-Flores L, Escamilla JE, Kennedy C. Indole-3-acetic acid biosynthesis is deficient in Gluconacetobacter diazotrophicus strains with mutations in cytochrome c biogenesis genes. J Bacteriol. 2004;186(16):5384-91. https://doi.org/10.1128/JB.186.16.5384-5391.2004 [ Links ]
11. Vieira PD, Motta CM, Lima D, Torres JB, Quecine MC, Azevedo JL, et al. Endophytic fungi associated with transgenic and non-transgenic cotton. Mycology. 2011;2(2):91-97. [ Links ]
12. Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol. 2005;51(2):215-229. https://doi.org/10.1016/j.femsec.2004.08.006 [ Links ]
13. Lowman S, Kim-Dura S, Mei C, Nowak J. Strategies for enhancement of switchgrass (Panicum virgatum L.) performance under limited nitrogen supply based on utilization of N-fixing bacterial endophytes. Plant Soil. 2016;405(1-2):47-63. https://doi.org/10.1007/s11104-015-2640-0 [ Links ]
14. Maida I, Chiellini C, Mengoni A, Bosi E, Firenzuoli F, Fondi M, et al. Antagonistic interactions between endophytic cultivable bacterial communities isolated from the medicinal plant Echinacea purpurea. Environ Microbiol. 2015;18:2357-2365. http://dx.doi.org/10.1111/1462-2920.12911 [ Links ]
15. Rijavec T, Lapanje A, Dermastia M, Rupnik M. Isolation of bacterial endophytes from germinated maize kernels. Can J Microbiol. 2007;53:802-808. https://doi.org/10.1139/W07-048 [ Links ]
16. Rodríguez-Blanco A, Sicardi M, Frioni L. Plant genotype and nitrogen fertilization effects on abundance and diversity of diazotrophic bacteria associated with maize (Zea mays L.). Biol Fert Soils. 2015;51(3):391-402. https://doi.org/10.1007/s00374-014-0986-8 [ Links ]
17. Araújo JM, Silva AC, Azevedo JL. Isolation of endophytic actinomycetes from roots and leaves of maize (Zea mays L.). Braz Arch Biol Technol. 2000;43(4):1-2. http://dx.doi.org/10.1590/S1516-89132000000400016 [ Links ]
18. Liu Y, Zuo S, Xu L, Zou Y, Song W. Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch Microbiol. 2012;194(12):1001-1012. http://dx.doi.org/10.1007/s00203-012-0836-8 [ Links ]
19. Roesch LFW, Camargo FAO, Bento FM, Triplett EW. Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil. 2008;302(1-2):91-104. http://dx.doi.org/10.1007/s11104-007-9458-3 [ Links ]
20. Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycol Res. 2007;99(2):185-206. http://dx.doi.org/10.1080/15572536.2007.11832578 [ Links ]
21. Güssow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res. 1989;17:4000 [ Links ]
22. Obi LU, Atagana HI, Adeleke RA. Isolation and characterisation of crude oil sludge degrading bacteria. SpringerPlus. 2016;5(1), Art. #1946, 13 pages. https://doi.org/10.1186/s40064-016-3617-z [ Links ]
23. Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695-700. [ Links ]
24. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95-98. [ Links ]
25. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(suppl 1):141-145. https://doi.org/10.1093/nar/gkn879 [ Links ]
26. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537-7541. https://doi.org/10.1128/AEM.01541-09 [ Links ]
27. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772-780. https://doi.org/10.1093/molbev/mst010 [ Links ]
28. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729. https://doi.org/10.1093/molbev/mst197 [ Links ]
29. Wilson M, Knight D. Methods of plant pathology. Tuite J, editor. London: Academic; 1952. [ Links ]
30. Rao NSS. Soil microorganisms and plant growth. New Delhi: Oxford and IBH Publishing Co; 1977. [ Links ]
31. Mwashasha RM, Hunja M, Tani A. Screening of bacterial and fungal isolates for their plant growth promoting activities. In: Proceedings of the 2012 JKUAT Scientific, Technological and Industrialization Conference; 2012 November 15-16; Nairobi, Kenya. Nairobi: Jomo Kenyatta University of Agriculture and Technology, Research Production and Extension Division; 2012. http://journals.jkuat.ac.ke/index.php/jscp/article/view/868 [ Links ]
32. Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170(1):265-270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x [ Links ]
33. Adeleke R, Cloete TE, Khasa DP. Culturable microorganisms associated with Sishen iron ore and their potential roles in biobeneficiation. World J Microbiol Biotechnol. 2012;28(3):1057-1070. https://doi.org/10.1007/s11274-011-0904-2 [ Links ]
34. Andreote FD, Azevedo JL, Araújo WL. Assessing the diversity of bacterial communities associated with plants. Braz J Microbiol. 2009;40:417-432. https://doi.org/10.1590/S1517-83822009000300001 [ Links ]
35. Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142-160. https://doi.org/10.1111/j.1574-6941.2007.00375.x [ Links ]
36. Ter Braak CJF, Smilauer P. CANOCO reference manual and CanoDraw for Windows user's guide: Software for canonical community ordination (Version 4.5). Ithaca, NY: Microcomputer Power; 2002. [ Links ]
37. Andreote FD, Rossetto PB, Mendes R, Avila LA, Labate CA, Pizzirani-Kleiner AA, et al. Bacterial community in the rhizosphere and rhizoplane of wild type and transgenic eucalyptus. World J Microbiol Biotechnol. 2009;25:1065-1073. https://doi.org/10.1007/s11274-009-9990-9 [ Links ]
38. Whittaker RH, Levin SA, Root RB. Niche, habitat, and ecotope. Amer Nat. 1973;107(955):321-338. http://doi.org/10.1086/282837 [ Links ]
39. Ter Braak CJ. Canonical correspondence analysis: A new eigenvector technique for multivariate direct gradient analysis. FEMS Microbiol Ecol. 1986;67(5):1167-1179. http://doi.org/10.2307/1938672 [ Links ]
40. James C. Global status of commercialized biotech/GM crops. ISAAA brief no.39. Ithaca, NY: ISAAA; 2010. [ Links ]
41. Saxena D, Flores S, Stotzky G. Bt toxin is released in root exudates from 12 transgenic corn hybrids rep resenting three transformation events. Soil Biol Biochem. 2002;34(1):133-137. http://doi.org/10.1016/S0038-0717(01)00161-4 [ Links ]
42. Pereira P, Ibáñez F, Rosenblueth M, Etcheverry M, Martínez-Romero E. Analysis of the bacterial diversity associated with the roots of maize (Zea mays L.) through culture-dependent and culture-independent methods. FEMS Microbiol Ecol. 2011;2011, Art. #938546, 10 pages. http://doi.org/10.5402/2011/938546 [ Links ]
43. Kumar DS, Hyde KD. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004;17:69-90. http://hdl.handle.net/10722/223074 [ Links ]
44. Montañez A, Blanco AR, Barlocco C, Beracochea M, Sicardi M. Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (Zea mays L.) and their inoculation effects in vitro. Appl Soil Ecol. 2012;58:21-28. http://doi.org/10.1016/j.apsoil.2012.02.009 [ Links ]
45. Doebereiner J, Pedrosa FO. Nitrogen-fixing bacteria in nonleguminous crop plants. New York: Science Tech; 1987. [ Links ]
46. Lynch JM. Microbial metabolites. In: Lynch JM, editor. The rhizosphere. Chichester: Wiley; 1990. p. 177-206. [ Links ]
47. Pandey RK, Maranville JW, Admou A. Deficit irrigation and nitrogen effects on maize in a Sahelian environment: I. Grain yield and yield components. Agric Water Manage. 2000;46(1):1-3. http://doi.org/10.1016/S0378-3774(00)00073-1. [ Links ]
48. Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7(11):847-859. http://dx.doi.org/10.1038/nrm2020 [ Links ]
49. Mashiane RA, Ezeokoli OT, Adeleke RA, Bezuidenhout CC. Metagenomic analyses of bacterial endophytes associated with the phyllosphere of a Bt maize cultivar and its isogenic parental line from South Africa. World J Microbiol Biotechnol. 2017;33(4):80. https://doi.org/10.1007/s11274-017-2249-y [ Links ]
 Correspondence:
Correspondence:
Rasheed A. Adeleke
Email: adeleker@arc.agric.za
Received: 19 Jan. 2017
Revised: 13 Apr. 2017
Accepted: 03 May 2018
Published: 30 July 2018
FUNDING: National Research Foundation (South Africa)
Supplementary Data
The supplementary data is available in pdf: [Supplementary data]














