Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.113 n.5-6 Pretoria May./Jun. 2017
http://dx.doi.org/10.17159/sajs.2017/20160287
RESEARCH ARTICLE
Isolation and characterisation of endocrine disruptor nonylphenol-using bacteria from South Africa
Lehlohonolo B. QhanyaI; Ntsane T. MthakathiI; Charlotte E. BoucherII; Samson S. MasheleI; Chrispian W. TheronII; Khajamohiddin SyedI
IUnit for Drug Discovery Research, Department of Health Sciences, Central University of Technology, Bloemfontein, South Africa
IIDepartment of Microbial, Biochemical and Food Biotechnology, University of the Free State, Bloemfontein, South Africa
ABSTRACT
Endocrine disrupting chemicals (EDCs) are synthetic chemicals that alter the function of endocrine systems in animals including humans. EDCs are considered priority pollutants and worldwide research is ongoing to develop bioremediation strategies to remove EDCs from the environment. An understanding of indigenous microorganisms is important to design efficient bioremediation strategies. However, much of the information available on EDCs has been generated from developed regions. Recent studies have revealed the presence of different EDCs in South African natural resources, but, to date, studies analysing the capabilities of microorganisms to utilise/degrade EDCs have not been reported from South Africa. Here, we report for the first time on the isolation and enrichment of six bacterial strains from six different soil samples collected from the Mpumalanga Province, which are capable of utilising EDC nonylphenol as a carbon source. Furthermore, we performed a preliminary characterisation of isolates concerning their phylogenetic identification and capabilities to degrade nonylphenol. Phylogenetic analysis using 16S rRNA gene sequencing revealed that four isolates belonged to Pseudomonas and the remaining two belonged to Enterobacteria and Stenotrophomonas. All six bacterial species showed degradation of nonylphenol in broth cultures, as HPLC analysis revealed 41-46% degradation of nonylphenol 12 h after addition. The results of this study represent the beginning of identification of microorganisms capable of degrading nonylphenol, and pave the way for further exploration of EDC-degrading microorganisms from South Africa.
SIGNIFICANCE:
• First report of endocrine disruptor nonylphenol-using bacteria from South Africa
• Six bacterial species capable of using nonylphenol as a carbon source were isolated
• Results will pave the way for further exploration of endocrine disruptors degrading microbes from South Africa
Keywords: biodegradation; endocrine disrupting chemicals; phylogenetic analysis; bioremediation; Pseudomonas
Introduction
Endocrine disruptors or endocrine disrupting chemicals (EDCs) are chemicals that can alter the functioning of endocrine systems in humans and other animals including wildlife, and can thus cause cancerous tumour development, birth defects and other developmental disorders.1,2 Many chemicals have been identified as EDCs, and many are used in the formulation of various pharmaceutical products, pesticides, industrial chemicals, heavy metals, persistent organochlorines and other organohalogens, alkylphenols, and synthetic and natural hormones.2,3, These environmental pollutants mimic natural hormones of the endocrine system and display either oestrogenic or androgenic activities.1,2,4 They can thus have adverse effects by either unnaturally inhibiting or stimulating the endocrine system and/or hormonal production.1,2,4 Exposure to EDCs increases the chance of physiological abnormalities and alters cognitive function in animals, including humans.1,2 Physiological abnormalities include low sperm count and decreased sperm quality5, as well as premature puberty in both girls6 and boys7. Several other metabolic disorders have been reported, including different types of cancers and thyroid-related problems including obesity.1,2,8
Investigations have also shown that these types of chemicals also affect other animals. Effects of EDCs on aquatic species have been well documented.9 EDCs have been reported to have adverse effects on invertebrates and wildlife populations.10 Female snails exposed to tributylin exhibited masculinisation (a disorder called imposex in which female snails develop a male sex organ, including a penis and vas deferens), which in turn led to a decline in the population.11 Alligators of Lake Apopka (Florida, USA) were reported to have impaired sexual development and function as a result of exposure to dichlorodiphenyltrichloroethane (DDT).12 Exposure to dichlorodiphenyldichloroethylene (DDE) resulted in a decline in numbers of bald eagles in Europe and North America.13
To date, information concerning EDCs has been primarily derived from studies conducted in developed countries.2 Much information is still, however, lacking from large parts of Africa, Asia and Central and South America.2 Studies on EDCs from South Africa in particular are very scarce. A report presented by the Water Research Commission of South Africa revealed the presence of EDCs in South African water.14 In addition to this report, studies conducted in a few places within South Africa have also revealed the presence of EDCs. DDT, DDE and phthalate esters have been found in Limpopo15-17; oestrone, oestradiol and oestriol (steroids hormones) in the Western Cape18 and in KwaZulu-Natal19; p-nonylphenol, diethylhexyl phthalate and dibutyl phthalate in Gauteng20; and lastly DDT, chlordane, hexachlorobenzene, heptachlor and endosulfan in the Eastern Cape21. In addition, a large number of EDCs was found in upstream and downstream sections of wastewater treatment plants.22,23
As a result of their adverse effects on humans and wildlife, EDCs are considered to be priority pollutants, and worldwide research is ongoing to develop remediation strategies to remove these chemicals from the environment. Strategies for removal - including advanced oxidation processes24, electrochemical separation and degradation technologies25 and bioremediation and combinatorial techniques26,27 - have been extensively investigated. Bioremediation is a particularly attractive approach, as it represents natural and economically feasible processes for detoxification of environmental pollutants under environmental conditions. An understanding of indigenous microorganisms is therefore important to facilitate the design of efficient bioremediation strategies. However, to date, studies on the analysis of the capabilities of microorganisms to utilise/degrade EDCs have not been reported from South Africa. This study is the first of its kind on the enrichment, isolation, identification and further assessment of the EDC-degradation capability of bacteria from South African soils.
Materials and methods
Soil sample collection and preparation
Soil samples were aseptically collected from soil at different coal-fired power stations in and around the Mpumalanga Province, South Africa. The selected sampling areas are represented in a schematic diagram with GPS coordinates (Figure 1). Soil samples (5 g) were re-suspended in 30 mL of DNase-free and RNase-free water.28,29 The samples were vigorously vortexed for 5 min, followed by incubation on a rotary shaker for 1 h at room temperature at 100 rpm.28,29 After incubation, the soil was allowed to settle out of solution (30 min), and the supernatants were collected and immediately used for isolation of microorganisms.
Medium preparation
All chemicals and reagents used in this study were purchased from Sigma-Aldrich (Johannesburg, South Africa), unless otherwise stated. Minimal medium28,29 with added trace element solution30 was used for isolation of microorganisms. The minimal medium consisted of 8.5 g/L Na2HPO4.2H2O, 3.0 g/L KH2PO4, 0.5 g/L NaCl, 1.0 g/L NH4Cl, 0.5 g/L MgSO4.7H2O, 14.2 mg/L CaCl2 and 0.15 g/L KCL. The minimal medium was supplemented with 10 mL of trace element solution30, consisting of 0.4 mg/L CuSO4, 1.0 mg/L KI, 4.0 mg/L MnSO4.H2O, 4.0 mg/L ZnSO4.7H2O, 5.0 mg/L H3BO3, 1.2 mg/L Na2MO4.2H2O and 2.0 mg/L FeCl3.6H2O, per litre of medium. Technical grade nonylphenol (catalogue number 290858) was added as a sole source of carbon to a final concentration of 5 mM.
Enrichment procedure
Supernatant (1 mL) from the soil samples was used to inoculate 100 mL of minimal medium in a 500-mL conical flask, supplemented with nonylphenol as the sole carbon source. A control was set up to contain medium and nonylphenol, without inoculation of soil samples. After 4 weeks of incubation at 37 °C at 100 rpm, 1 mL of culture was used to inoculate fresh minimal medium (100 mL) with nonylphenol as the sole carbon source. This serial enrichment of bacterial isolates was repeated until a single, homogenous culture was obtained. Aliquots (100 µL) of cultures were spread on minimal medium agar plates with nonylphenol (5 mM) as the sole carbon source, to monitor the growth of microorganisms at 37 °C. The minimal medium plates with nonylphenol were prepared as described elsewhere.31 Bacterial growth was also analysed by measuring the absorbance at 600 nm.
Isolation of genomic DNA and amplification of 16S rRNA gene
Genomic DNA (gDNA) from bacterial isolates was extracted using the ZR Fungal/Bacterial DNA MiniPrep kit (catalogue number D6005, Inqaba Biotec, Pretoria, South Africa) according to the manufacturer's protocol. The gDNA was visualised using agarose gel electrophoresis, and gDNA concentration was measured using a SimpliNano microvolume spectrophotometer (catalogue number GE29-0617-12, Sigma-Aldrich, St. Louis, MO, USA). The isolated gDNA was used for amplification of the 16S rRNA gene. The 16S rRNA gene was amplified by polymerase chain reaction (PCR) using primers 63f and 1387r as described elsewhere.32 A KAPA HiFi HotStart PCR kit (catalogue number KK2501, KAPA Biosystems, Wilmington, MA, USA) was used to amplify the 16S rRNA gene according to manufacturer's instructions. The PCR products were run on a 0.8% agarose gel and were purified using the Wizard® SV Gel and PCR Clean-Up System (catalogue number A9281, Promega, Madison, WI, USA).
16S rRNA gene sequencing
Samples were prepared for sequencing using the BigDye™ Terminator V3.1 Cycle Sequencing Kit (catalogue number 4337455, Thermo Fischer Scientific, Waltham, MA, USA). The aforementioned primers 63f and 1387r32 were used for sequencing. The sequencing reactions were performed according to the parameters described by the manufacturer. Sequencing reactions were purified using the EDTA-ethanol method described by the manufacturer, and submitted for sequencing using a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Consensus sequences were derived from the sequences obtained from the forward and reverse primer reactions for each product, using Geneious® R9 9.1.2. software.
Phylogenetic analysis
16S rRNA gene sequences of bacterial isolates were subjected to BLAST analysis at NCBI (the US National Center for Biotechnology Information) against 16S ribosomal RNA sequences (Bacteria and Archaea) to identify the closest homologs. Among the resulting hits, the 16S rRNA sequences with 100% or 99% identity homologs were selected. Based on the obtained bacterial species, the type strains belonging to each species were selected, and the 16S rRNA sequences were retrieved from elsewhere (http://www.bacterio.net/). The Escherichia coli ATCC 11775 type strain 16S rRNA gene sequence (also retrieved from http://www.bacterio.net/) was used as an out-group. Phylogenetic analysis was carried out using the maximum likelihood method based on the Tamura-Nei model.33 Initial tree(s) for the heuristic search were obtained by applying the neighbour-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood approach. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5.34 Phylogenetic analysis included the isolate 16S rRNA gene sequence, hit homologs and type strain 16S rRNA gene sequences. The phylogenetic tree was presented with branch lengths, and the bacterial isolates identified in this study are highlighted in bold font.
Nonylphenol degradation
A degradation study using whole cells was carried out as described elsewhere35,36 to assess the capabilities of the bacterial isolates to degrade nonylphenol. A single colony of isolates from minimal medium plate containing nonylphenol as the carbon source was used to inoculate 5 mL of Luria-Bertani broth, which was then cultured overnight at 150 rpm at 37 °C. The growth of the isolates was measured at 600 nm after diluting the culture in Luria-Bertani broth. The cultures were then washed twice with saline (0.9% sodium chloride solution), followed by inoculation with an equal amount of each overnight bacterial culture for all six isolates onto separate, fresh minimal media (5 mL) containing nonylphenol (2.5 mM) as a carbon source in 50-mL glass tubes (test cultures). The test cultures were incubated for 12 h at 37 °C at 150 rpm. After incubation, 5 mL of ethyl acetate was added to the test cultures, which were then vortexed for 5 min at maximum speed, followed by centrifugation for 5 min at 2500 g at room temperature. After centrifugation, two distinct fractions were separated by a thin middle layer composed of bacterial cell debris. The upper organic fraction containing nonylphenol was removed from the lower aqueous fraction into a fresh glass tube. The extraction was repeated twice, followed by evaporation of the organic fraction. The remaining residue was re-suspended in 200 µL of HPLC-grade methanol. Minimal medium with nonylphenol but without culture was used as a control and treated the same as the test culture.
HPLC analysis of nonylphenol was carried out following the method described elsewhere, with modifications.35,36 Briefly, the abovementioned methanol samples were filtered through 0.45-µm glass fibre filters and analysed using a Shimadzu Prominence instrument (Shimadzu, Roodepoort, South Africa) equipped with a C18 analytical column (4.6 mm×250 mm; particle size 5 µm from Sigma-Aldrich, South Africa) and with a dual wavelength UV/Vis detector. Separation was achieved using a 22.5-min linear gradient of acetonitrile in water (50% to 96.5%, and then re-equilibrated for 10 min at 50% acetonitrile at a flow rate of 1.25 mL/min). A volume of 5 µL of sample was injected for analysis. Nonylphenol was detected at 277 nm, and the percentage degradation of nonylphenol by test cultures was related to the control nonylphenol, which was taken as 100%.
Statistical analysis
All experiments were carried out in triplicate and results were subjected to statistical analysis as described elsewhere.35,36 The activities, in terms of percentage degradation, of the different bacterial isolates were analysed for means and standard deviations and compared for statistical differences using a Student's t-test on GraphPad QuickCalcs software package (GraphPad Software Inc., CA, USA).
16S rRNA gene sequences accession numbers
16S rRNA gene sequences of bacterial isolates identified in this study were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/), with the following accession numbers: KX364074 (Pseudomonas nitroreducens strain LBQSKN1), KX364075 (Pseudomonas putida strain LBQSKN2), KX364076 (Stenotrophomonas sp. LBQSKN3), KX364077 (Enterobacter asburiae strain LBQSKN4), KX364078 (Pseudomonas sp. LBQSKN5) and KX364079 (Pseudomonas sp. LBQSKN6).
Results and discussion
Enrichment and isolation of nonylphenol-utilising bacteria
The sampling areas selected for this study (represented in Figure 1) have been reported to harbour polycyclic aromatic hydrocarbons (PAHs).37 PAHs are hydrophobic compounds well known for their carcinogenicity and mutagenicity towards humans.38,39 In this study, we aimed to test the ability of bacterial species growing in the presence of PAHs to degrade EDCs, as these chemicals are also hydrophobic and aromatic in nature. To isolate microorganisms capable of utilising nonylphenol as a sole source of carbon, we followed a standard enrichment method. Soil samples collected from six different places (Figure 1) were inoculated into minimal medium supplemented with nonylphenol as a carbon source. After 4 weeks of incubation, growth of bacteria was observed on minimal medium plates supplemented with nonylphenol as a carbon source, as well as assessed through spectrophotometry. The initial bacterial growth on plates was non-homogenous, suggesting the presence of more than one type of species. After three successive serial cultures, a homogenous population of bacteria was observed on minimal medium plates, indicating that successive serial culturing resulted in the enrichment of a single type of bacteria that are capable of utilising nonylphenol as a sole source of carbon. In this study, six bacteria were isolated from the six different soil samples.
Identification of bacterial isolates
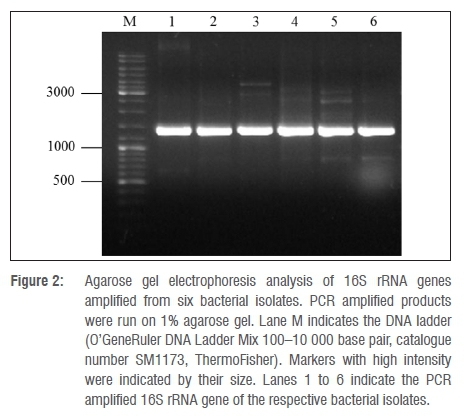
In order to identify the enriched bacterial isolates, 16S rRNA gene sequence-based phylogenetic analysis was carried out. The 16S rRNA genes from the gDNA of bacterial isolates were PCR amplified using the 63f and 1387r primer set as described elsewhere.32 Analysis of the PCR amplified products on agarose gel showed prominent DNA bands with approximate sizes of ≥1200 base pairs (Figure 2). This analysis indicates specific amplification of the 16S rRNA gene. The amplified 16S rRNA gene was gel purified and subjected to sequence analysis using the same primers used for its amplification. Sequence analysis was performed using both forward and reverse primers, yielding a consensus sequence of 300-500 overlapping base pairs between the sequences. The sizes of the 16S rRNA sequences obtained for each of the bacterial isolates are presented in Table 1. The 16S rRNA sequence of Isolates 1 and 2 showed 100% identity to Pseudomonas spp., while Isolates 5 and 6 also had 99% identity to Pseudomonas spp. (Table 1). Isolate 3 showed 99% identity to Stenotrophomonas spp. and Isolate 4 showed 99% identity to Enterobacter spp. This indicates that most of the isolates belong to Pseudomonas (Table 1). Phylogenetic analysis of isolates based on 16S rRNA gene sequences compared to the 16S rRNA gene sequences of hit species, highlighted the differential alignment of bacterial isolates with different species (Figure 3). Based on the phylogenetic alignment, the six bacterial isolates were named as shown in Table 1. Furthermore, homology analysis (per cent identity) of 16S rRNA gene sequences among bacterial isolates (Table 2) revealed that Isolates 3 and 4 have low per cent identity compared with that of the other isolates, clearly reinforcing that they in fact belong to different bacterial genera. Species assigned to Pseudomonas on the other hand showed high per cent identity (Table 2), demonstrating that they belong to the same genus.
Degradation of nonylphenol by bacterial species
Whole-cell nonylphenol degradation experiments were carried out to assess the nonylphenol degradation capability of each bacterial isolate. As shown in Figure 4, all bacterial isolates showed degradation of nonylphenol. The degradation of nonylphenol by bacterial isolates ranged from 41% to 46% (Figure 4). However, the difference in percentage of nonylphenol degradation by all six bacterial species was considered to be the same, because the percentage differences among the isolates was not statistically significant (0.2<p<0.7). Nonylphenol degradation by the bacterial species identified in this study is reinforced by the literature. Species belonging to the genus Pseudomonas have been shown to degrade EDCs such as di-n-butyl phthalate40, p-nonylphenol41 and polyethoxylated nonylphenols42,43. Bacterial species belonging to Stenotrophomonas were previously found to be capable of using either nonylphenol or octylphenol as a sole carbon source.44 For species belonging to the well-known human-pathogenic and plant association Enterobacter, degradation of EDCs has been reported particularly for bisphenol A45, polychlorinated biphenyls46, endosulfan47, dibutyl phthalate48 and nonylphenol49.

All of the bacterial species isolated in this study also have the capability to degrade PAHs. PAH degradation by Pseudomonas species is well reported.50-54 Degradation of PAHs using Stenotrophomonas55, in particular Stenotrophomonas maltophilia56-58, has been investigated. Hydrocarbon degradation capabilities for some of these species have also been demonstrated with aliphatic59 and aromatic hydrocarbons60. This suggests that the soil samples used in this study, from areas where PAHs were reported to be present, harbour bacterial species that are capable of degrading both classes of xenobiotics, PAHs and EDCs.
Conclusion
The distribution of EDCs, their effects towards living organisms and microorganisms capable of degrading ECDs, and the mechanisms of EDC degradation have been thoroughly documented by the developed world. Information on these matters is, however, lacking from Africa, Asia and Central and South America.
Our study is thus the first of its kind from South Africa, in which we successfully enriched, isolated, identified and demonstrated nonylphenol degradation capabilities of indigenous bacterial strains. The areas from which soil samples were collected were previously reported to be polluted with PAHs, and their selection resulted in the isolation of bacterial species capable of degrading EDC nonylphenol, suggesting that these organisms have the capability to degrade a variety of xenobiotic chemicals. Further investigations on the capacity of the isolates to degrade different EDCs and PAHs are currently underway. The results presented in this study will lead to the isolation and characterisation of microorganisms from different parts of South Africa that are capable of degrading different EDCs, and will thus enrich EDC-related information from Africa.
Acknowledgements
We thank Sarel Marais at the Department of Biotechnology, University of the Free State for technical assistance with HPLC analyses. K.S. and S.S.M. express their sincere gratitude to the Central University of Technology (Bloemfontein, South Africa) for a grant from the University Research and Innovation Fund and Emerging Researcher Award (to K.S.). K.S. and S.S.M. thank the Department of Higher Education and Training (South Africa) and the Technology Innovation Agency (South Africa) for a research grant. S.S.M., C.W.T. and C.E.B. also thank the National Research Foundation (South Africa) for a research grant. L.B.Q. and N.T.M. are thankful to the National Research Foundation and Central University of Technology for master's fellowships.
Authors' contributions
K.S. and C.W.T. conceived and designed the experiments. K.S. provided funding for the study. All authors were involved in performing the experiments, analysing the data and writing the manuscript.
References
1. Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, et al. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr Rev. 2009;30(4):293-342. https://doi.org/10.1210/er.2009-0002 [ Links ]
2. World Health Organization (WHO) / United Nations Environment Programme (UNEP). State of the science of endocrine disrupting chemicals - 2012 In: Bergman A, Heindel JJ, Jobling S, Kidd KA, Zoeller RT, editors. Geneva: UNEP / Inter-Organization Programme for the Sound Management of Chemicals, WHO; 2013. Available from: http://www.who.int/ceh/publications/endocrine/en/ [ Links ]
3. International Panel of Chemical Pollution (IPCP). Overview report I: A compilation of lists of chemicals recognised as endocrine disrupting chemicals (EDCs) or suggested as potential EDCs [document on the Internet]. c2016 [cited 2016 Sep 12]. Available from: http://wedocs.unep.org/handle/20.500.11822/12218 [ Links ]
4. De Falco M, Forte M, Laforgia V. Estrogenic and anti-androgenic endocrine disrupting chemicals and their impact on the male reproductive system. Front Environ Sci. 2015;3, Art. #3, 12 pages. http://dx.doi.org/10.3389/fenvs.2015.00003 [ Links ]
5. Marques-Pinto A, Carvalho D. Human infertility: Are endocrine disruptors to blame? Endocr Connect. 2013;2(3):15-29. https://doi.org/10.1530/EC-13-0036 [ Links ]
6. Fowler PA, Bellingham M, Sinclair KD, Evans NP, Pocar P, Fischer B, et al. Impact of endocrine-disrupting compounds (EDCs) on female reproductive health. Mol Cell Endocrinol. 2012;355(2):231-239. https://doi.org/10.1016/j.mce.2011.10.021 [ Links ]
7. Zawatski W, Lee M. Male pubertal development: Are endocrine-disrupting compounds shifting the norms? J Endocrinol. 2013;218(2):1-12. https://doi.org/10.1530/JOE-12-0449 [ Links ]
8. Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine disrupting chemicals and disease susceptibility. J Steroid Biochem Mol Biol. 2011;127(3-5):204-215. https://doi.org/10.1016/j.jsbmb.2011.08.007 [ Links ]
9. Soares A, Guieysse B, Jefferson B, Cartmell E, Lester JN. Nonylphenol in the environment: A critical review on occurrence, fate, toxicity and treatment in wastewaters. Environ Int. 2008;34(7):1033-1049. https://doi.org/10.1016/j.envint.2008.01.004 [ Links ]
10. Flint S, Markle T, Thompson S, Wallace E. Bisphenol A exposure, effects, and policy: A wildlife perspective. J Environ Manage. 2012;104:19-34. https://doi.org/10.1016/j.jenvman.2012.03.021 [ Links ]
11. Matthiessen P, Gibbs PE. Critical appraisal of the evidence for tributyltin-mediated endocrine disruption in mollusks. Environ Toxicol Chem. 1998;17(1):37-43. https://doi.org/10.1002/etc.5620170106 [ Links ]
12. Guillette Jr LJ, Gross TS, Masson GR, Matter JM, Percival HF, Woodward AR. Developmental abnormalities of the gonads and abnormal sex hormone concentrations in juvenile alligators from contaminated and control lakes in Florida. Environ Health Perspect. 1994;102(8):680-688. https://doi.org/10.2307/3432198 [ Links ]
13. Cooke AS. Shell thinning in avian eggs by environmental pollutants. Environ Pollut. 1973;4(2):85-152. http://dx.doi.org/10.1016/0013-9327(73)90009-8 [ Links ]
14. Burger AEC, Nel A. Scoping study to determine the potential impact of agricultural chemical substances (pesticides) with endocrine disruptor properties on water resources of South Africa. WRC report no. 1774/1/08. Pretoria: Water Research Commission; 2008. [ Links ]
15. Aneck-Hahn NH, Schulenburg GW, Bornman MS, Farias P, De Jager C. Impaired semen quality associated with environmental DDT exposure in young men living in a malaria area in the Limpopo Province, South Africa. J Androl. 2007;28(3):423-434. https://doi.org/10.2164/jandrol.106.001701 [ Links ]
16. Aneck-Hahn NH, Bornman MS, De Jager C. Oestrogenic activity in drinking waters from a rural area in the Waterberg District, Limpopo Province, South Africa. Water SA. 2009;35(3):245-252. [ Links ]
17. Fatoki OS, Bornman M, Ravandhalala L, Chimuka L, Genthe B, Adeniyi A. Phthalate ester plasticizers in freshwater systems of Venda, South Africa and potential health effects. Water SA. 2010;36(1):117-126. https://doi.org/10.4314/wsa.v36i1.50916 [ Links ]
18. Swart N, Pool E. Rapid detection of selected steroid hormones from sewage effluents using an ELISA in the Kuils River water catchment area, South Africa. J Immunoassay Immunochem. 2007;28(4):395-408. https://doi.org/10.1080/15321810701603799 [ Links ]
19. Manickum T, John W. Occurrence, fate and environmental risk assessment of endocrine disrupting compounds at the wastewater treatment works in Pietermaritzburg (South Africa). Sci Total Environ. 2014;468-469:584-597. https://doi.org/10.1016/j.scitotenv.2013.08.041 [ Links ]
20. Mahomed SI, Voyi KVV, Aneck-Hahn NH, De Jager C. Oestrogenicity and chemical target analysis of water from small-sized industries in Pretoria, South Africa. Water SA. 2008;34(3):357-364. [ Links ]
21. Fatoki OS, Awofolu OR. Levels of organochlorine pesticide residues in marine-, surface-, ground- and drinking waters from the Eastern Cape Province of South Africa. J Environ Sci Health. 2004;39(1):101-114. https://doi.org/10.1081/PFC-120027442 [ Links ]
22. Olujimi OO, Fatoki OS, Odendaal JP, Okonkwo JO. Endocrine disrupting chemicals (phenol and phthalates) in the South African environment: A need for more monitoring. Water SA. 2010;36(5):671-682. https://doi.org/10.4314/wsa.v36i5.62001 [ Links ]
23. Olujimi OO, Fatoki OS, Odendaal JP, Daso AP. Chemical monitoring and temporal variation in levels of endocrine disrupting chemicals (priority phenols and phthalate esters) from selected wastewater treatment plant and freshwater systems in Republic of South Africa. Microchem J. 2012,101:11-23. https://doi.org/10.1016/j.microc.2011.09.011 [ Links ]
24. Matilainen A, Sillanpää M. Removal of natural organic matter from drinking water by advanced oxidation processes. Chemosphere. 2010;80(4):351-365. https://doi.org/10.1016/j.chemosphere.2010.04.067 [ Links ]
25. Sirés I, Brillas E. Remediation of water pollution caused by pharmaceutical residues based on electrochemical separation and degradation technologies: A review. Environ Int. 2012;40:212-229. https://doi.org/10.1016/j.envint.2011.07.012 [ Links ]
26. Liu Z, Kanjo Y, Mizutani S. Removal mechanisms for endocrine disrupting compounds (EDCs) in wastewater treatment - Physical means, biodegradation, and chemical advanced oxidation: A review. Sci Total Environ. 2009;407(2):731-748. https://doi.org/10.1016/j.scitotenv.2008.08.039 [ Links ]
27. Gavrilescu M, Demnerova K, Aamand J, Agathos S, Fava F. Emerging pollutants in the environment: Present and future challenges in biomonitoring, ecological risks and bioremediation. Nat Biotechnol. 2015;32(1):147-156. https://doi.org/10.1016/j.nbt.2014.01.001 [ Links ]
28. Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59(6):1927-1930. [ Links ]
29. Zhang H, Kallimanis A, Koukkou AI, Drainas C. Isolation and characterization of novel bacteria degrading polycyclic aromatic hydrocarbons from polluted Greek soils. Appl Microbiol Biotechnol. 2004;65(1):124-131. https://doi.org/10.1007/s00253-004-1614-6 [ Links ]
30. Zeng J, Lin XG, Zhang J, Li XZ. Isolation of polycyclic aromatic hydrocarbons (PAHs)-degrading Mycobacterium spp. and the degradation in soil. J Hazard Mater. 2010;183(1-3):718-723. https://doi.org/10.1016/j.jhazmat.2010.07.085 [ Links ]
31. Porter AW, Hay AG. Identification of opdA, a gene involved in biodegradation of the endocrine disrupter octylphenol. Appl Environ Microbiol. 2007;73(22):7373-7379. https://doi.org/10.1128/AEM.01478-07 [ Links ]
32. Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64(2):795-799. [ Links ]
33. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512-526. [ Links ]
34. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-2739. https://doi.org/10.1093/molbev/msr121 [ Links ]
35. Syed K, Porollo A, Lam YW, Yadav JS. A fungal P450 (CYP5136A3) capable of oxidizing polycyclic aromatic hydrocarbons and endocrine disrupting alkylphenols: Role of Trp129 and Leu324. PLoS ONE. 2011;6(12), e28286, 14 pages. https://doi.org/10.1371/journal.pone.0028286. [ Links ]
36. Syed K, Porollo A, Lam YW, Grimmet PE, Yadav JS. A catalytically versatile fungal P450 monooxygenase (CYP63A2) capable of oxidizing higher polycyclic aromatic hydrocarbons, alkylphenols and alkanes. Appl Environ Microbiol. 2013;79(8):2692-2702. https://doi.org/10.1128/AEM.03767-12 [ Links ]
37. Okedeyi OO, Nindi MM, Dube S, Awofolu OR. Distribution and potential sources of polycyclic aromatic hydrocarbons in soils around coal-fired power plants in South Africa. Environ Monti Assess. 2012;185(3):2073-2082. https://doi.org/10.1007/s10661-012-2689-7 [ Links ]
38. Pashin YV, Bakhitova LM. Mutagenic and carcinogenic properties of polycyclic aromatic hydrocarbons. Environ Health Perspect. 1979;30:185-189. [ Links ]
39. Rengarajan T, Rajendran P, Nandakumar N, Lokeshkumar B, Rajendran P, Nishigaki I. Exposure to polycyclic aromatic hydrocarbons with special focus on cancer. Asian Pac J Trop Biomed. 2015;5(3):182-189. http://doi.org/10.1016/S2221-1691(15)30003-4 [ Links ]
40. Liao CS, Chen LC, Chen BS, Lin SH. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus radiodurans and Pseudomonas stutzeri. Chemosphere. 2010;78(3):342-346. https://doi.org/10.1016/j.chemosphere.2009.10.020 [ Links ]
41. Chakraborty J, Dutta TK. Isolation of a Pseudomonas sp. capable of utilizing 4-nonylphenol in the presence of phenol. J Microbiol Biotechnol. 2006;16(11):1740-1746. [ Links ]
42. John DM, White GF. Mechanism for biotransformation of nonylphenol polyethoxylates to xenoestrogens in Pseudomonas putida. J Bacteriol. 1998;180(17):4332-4338. [ Links ]
43. Ruiz Y, Medina L, Borusiak M, Ramos N, Pinto G, Valbuena O. Biodegradation of polyethoxylated nonylphenols. ISRN Microbiol. 2013;2013, Art. #284950, 9 pages. http://dx.doi.org/10.1155/2013/284950. [ Links ]
44. Toyama T, Murashita M, Kobayashi K, Kikuchi S, Sei K, Tanaka Y, et al. Acceleration of nonylphenol and 4-tert-octylphenol degradation in sediment by Phragmites australis and associated rhizosphere bacteria. Environ Sci Technol. 2011;45(15):6524-6530. https://doi.org/10.1021/es201061a [ Links ]
45. Badiefar l, Yakhchali B, Rodriguez-Couto S, Veloso A, Garcia-Arenzana J-M, Matsumura Y, et al. Biodegradation of bisphenol A by the newly-isolated Enterobacter gergoviae strain BYK-7 enhanced using genetic manipulation. RSC Advances. 2015;5(37):29563-29572. https://doi.org/10.1039/C5RA01818H. [ Links ]
46. Jia LY, Zheng AP, Xu L, Huang XD, Zhang Q, Yang FL. Isolation and characterization of comprehensive polychlorinated biphenyl degrading bacterium, Enterobacter sp. LY402. J Microbiol Biotechnol. 2008;18(5):952-957. [ Links ]
47. Abraham J, Silambarasan S. Plant growth promoting bacteria Enterobacter asburiae JAS5 and Enterobacter cloacae JAS7 in mineralization of endosulfan. Appl Biochem Biotechnol. 2015;175(7):3336-3348. https://doi.org/10.1007/s12010-015-1504-7 [ Links ]
48. Fang CR, Yao J, Zheng YG, Jiang CJ, Hu LF, Wu YY, et al. Dibutyl phthalate degradation by Enterobacter sp. T5 isolated from municipal solid waste in landfill bioreactor. Int Biodeterior Biodegradation. 2010;64(6):442-446. https://doi.org/10.1016/j.ibiod.2010.04.010 [ Links ]
49. Kageyama S, Morooka N. Effect of detergents on growth of bacteria. J Home Economics Japan. 2005;56(1):41-47. [ Links ]
50. Dong C, Bai X, Sheng H, Jiao L, Zhou H, Shao Z. Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences. 2015;12(7):2163-2177. https://doi.org/10.5194/bg-12-2163-2015 [ Links ]
51. Ma Y, Wang L, Shao Z. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol. 2006;8(3):455-465. https://doi.org/10.1111/j.1462-2920.2005.00911.x [ Links ]
52. Ma J, Xu L, Jia L. Degradation of polycyclic aromatic hydrocarbons by Pseudomonas sp. JM2 isolated from active sewage sludge of chemical plant. J Environ Sci. 2012;24(12):2141-2148. https://doi.org/10.1016/S1001-0742(11)61064-4 [ Links ]
53. Ma J, Xu L, Jia L. Characterization of pyrene degradation by Pseudomonas sp. strain Jpyr-1 isolated from active sewage sludge. Bioresour Technol. 2013;140:15-21. https://doi.org/10.1016/j.biortech.2013.03.184 [ Links ]
54. Zhang Z, Hou Z, Yang C, Ma C, Tao F, Xu P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour Technol. 2011;102(5):4111-4116. https://doi.org/10.1016/j.biortech.2010.12.064 [ Links ]
55. Mangwani N, Shukla SK, Kumari S, Rao TS, Das S. Characterization of Stenotrophomonas acidaminiphila NCW-702 biofilm for implication in the degradation of polycyclic aromatic hydrocarbons. J Appl Microbiol. 2014;117(4):1012-1024. https://doi.org/10.1111/jam.12602 [ Links ]
56. Boonchan S, Britz ML, Stanley GA. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol Bioeng. 1998;59(4):482-494. https://doi.org/10.1002/(SICI)1097-0290(19980820)59:4<482::AID-BIT11>3.0.CO;2-C [ Links ]
57. Juhasz AL, Stanley GA, Britz ML. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10,003. Lett Appl Microbiol. 2000;30(5):396-401. https://doi.org/10.1046/j.1472-765x.2000.00733.x [ Links ]
58. Singh A, Kumar K, Pandey AK, Sharma A, Singh SB, Kumar K, et al. Pyrene degradation by biosurfactant producing bacterium Stenotrophomonas maltophilia. Agric Res. 2015;4(1):42-47. https://doi.org/10.1007/s40003-014-0144-4 [ Links ]
59. Yousaf S, Ripka K, Reichenauer TG, Andria V, Afzal M, Sessitsch A. Hydrocarbon degradation and plant colonization by selected bacterial strains isolated from Italian ryegrass and birdsfoot trefoil. J Appl Microbiol. 2010;109(4):1389-1401. https://doi.org/10.1111/j.1365-2672.2010.04768.x [ Links ]
60. Obayori OS, Salam LB, Omotoyo IM. Degradation of weathered crude oil (Escravos Light) by bacterial strains from hydrocarbons-polluted site. Afr J Microbiol Res. 2012;6(26):5426-5432. [ Links ]
 Correspondence:
Correspondence:
Khajamohiddin Syed
khajamohiddinsyed@gmail.com
Received: 21 Sep. 2016
Revised: 28 Nov. 2016
Accepted: 05 Dec. 2016
FUNDING: Central University of Technology; National Research Foundation (South Africa); Department of Higher Education and Training (South Africa); Technology and Innovation Agency (South Africa)














