Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.112 n.9-10 Pretoria Sep./Oct. 2016
http://dx.doi.org/10.17159/sajs.2016/20160106
REVIEW ARTICLES
http://dx.doi.org/10.17159/sajs.2016/20160106
New light on vitamin B12: The adenosylcobalamin-dependent photoreceptor protein CarH
Susan M. Chemaly
School of Chemistry, University of the Witwatersrand, Johannesburg, South Africa
ABSTRACT
Adenosylcobalamin (AdoCbl), or coenzyme B12, is a cofactor for enzymes important in metabolism in humans (and other mammals) and bacteria. AdoCbl contains a Co-C bond and is extremely light sensitive, but, until recently, this light sensitivity appeared to have no physiological function. Recently, AdoCbl has been found to act as cofactor for a photoreceptor protein (CarH) that controls the expression of DNA coding for transcription of the proteins needed for synthesis of carotenes in certain non-photosynthetic bacteria. In 2015, the X-ray crystal structures of two dark states of the photoreceptor protein from the bacterium Thermus thermophilus were determined: CarH bound to AdoCbl and CarH bound to a large portion of the cognate DNA operator (and AdoCbl); a light state was also determined in which CarH was bound to cobalamin in which the Co-C bond had been broken. The breaking of the Co-C bond of Ado-Cbl acts as a trigger for the regulatory switch that allows the transcription of DNA. In the two dark states AdoCbl is bound to a conserved histidine from CarH, which displaces the lower 5,6-dimethylbenzimidazole ligand of AdoCbl. In the light state the 5'-deoxyadenosyl group of AdoCbl is replaced by a second histidine from CarH, giving a bis-histidine cobalamin and 4',5'-anhydroadenosine. Genes for B12-dependent photoreceptors are widespread in bacteria. Control of DNA transcription may represent an evolutionarily ancient function of AdoCbl, possibly pre-dating its function as a protein cofactor.
SIGNIFICANCE:
• A new function for adenosylcobalamin, a light-sensitive form of vitamin B12 with a Co-C bond, has been discovered in bacteria
• Some non-photosynthetic bacteria use adenosylcobalamin as a cofactor for the protein CarH, which controls DNA transcription
• Three X-ray crystal structures of CarH have been determined: bound to adenosylcobalamin, DNA and after light exposure
• A mechanism of action for CarH, based on its structure and on model reactions of vitamin B12, is proposed
Keywords: photolysis; coenzyme B12; light-sensing protein; DNA transcription; X-ray crystallography
Introduction
It is appropriate that in 2015, the UNESCO year of light, the X-ray crystal structure of a novel coenzyme B12-dependent photoreceptor protein was reported.1 Vitamin B12 was first discovered as cyanocobalamin (CNCbl), an inactive form of the vitamin, as an artefact of the isolation procedure, which utilised cyanide. (For reviews on vitamin B12, see 2-5). The coenzyme forms of vitamin B12 - adenosylcobalamin (AdoCbl) and methylcobalamin (MeCbl) (Figure 1) - are extremely unstable to light and were converted to CNCbl by photolysis during the work-up procedure of their original isolation. This sensitivity to light is considered to be a great nuisance by researchers because all experiments involving coenzymes must be performed in the dark or under dim red light. However, until recently, the photosensitivity of B12 coenzymes appeared to have no physiological function.1,6,7
Humans have only two vitamin B12-dependent enzymes: methylmalonyl-coenzyme A (CoA) mutase which utilises AdoCbl as its coenzyme and methionine synthase which utilises MeCbl. Methylmalonyl-CoA mutase, an example of an isomerase enzyme, converts methylmalonyl-CoA to succinyl-CoA in the oxidation of odd-chain fatty acids from the degradation of isoleucine and valine. Methionine synthase, an example of a methyl transfer enzyme, converts homocysteine to methionine. Humans (and other mammals) are unable to synthesise cobalamins and rely on dietary sources for this essential vitamin. Bacteria are much more versatile and some can synthesise cobalamins de novo as well as use them in a much wider variety of enzyme reactions. Bacteria utilise MeCbl in the fixation of carbon dioxide through the acetyl-CoA pathway, methanogenesis and in a variety of methylation reactions, including some involved in the synthesis of the corrinoid precursors of cobalamins. AdoCbl-requiring enzymes in bacteria include several isomerases, amino mutases, diol dehydratase, ethanolamine ammonia lyase and a ribonucleotide reductase.
The mechanism of action of AdoCbl-requiring enzymes involves homolysis of the Co-C bond of AdoCbl to give cobalamin in the Co(II) oxidation state and the 5'-deoxyadenosyl free radical. This reaction is similar to that taking place when free AdoCbl is photolysed. For AdoCbl-requiring enzymes, the 5'-deoxyadenosyl free radical takes part in rearrangement of substrate to product. For photolysis of free AdoCbl in the absence of oxygen, the 5'-deoxyadenosyl radical cyclises to give 5',8-cycloadenosine8 and in the presence of oxygen it gives adenosine 5'-aldehyde and 5'-peroxyadenosine (Scheme 1)9-11.
Dorothy Hodgkin determined the X-ray crystal structure of vitamin B12 in 1955.12 This structure was part of the research for which she was awarded a Nobel prize in chemistry. The structures of a large number of cobalamins, including AdoCbl and MeCbl, were subsequently determined. This determination was followed, from the mid-1990s, by the protein X-ray crystal structures of first the MeCbl-binding subunit of methionine synthase13 and then a large variety of other cobalamin-dependent enzymes, including methylmalonyl-CoA mutase14.
X-ray crystallography has shown that B12 enzymes can bind AdoCbl in two ways: in the 'base-off' form whereby a histidine from the protein displaces the 5,6-dimethylbenzimidazole (dbzm) base, as in methionine synthase13 and methylmalonylCoA mutase14 and in a 'base-on' form as in diol dehydratase and ethanolamine ammonia lyase5. X-ray crystallography has been invaluable in providing insight into the structures and reaction mechanisms of B12 enzymes.
AdoCbl is also known to interact directly with messenger RNA in ribo-switches. (A riboswitch is a mRNA which interacts directly and selectively with a small molecule.15) One of the first riboswitches to be discovered was the E. coli btuB mRNA,16 which interacts selectively with AdoCbl in order to control the synthesis of the BtuB transmembrane protein, which transports corrinoids across the outer membrane of the bacteria. At low AdoCbl concentrations the riboswitch is 'on', and at high concentrations the switch is 'off'. In the 'off' position, AdoCbl binds to the mRNA, changing its three-dimensional structure, preventing the association of mRNA with the ribosome and stopping the synthesis of BtuB. AdoCbl riboswitches are widespread in bacteria and are involved in the synthesis and transport of cobalamins and the transport of metal ions.16,17
Adenosylcobalamin-dependent photoreceptor proteins in bacteria
Recently, a new group of photoreceptor proteins that uses AdoCbl to sense light was discovered in certain non-photosynthetic bacteria.6,18-27 These bacteria produce carotenoids - yellow, orange or red pigments - which protect them from photo-oxidative damage by quenching the singlet oxygen free radicals produced by absorption of energy from light.19 The photoreceptor proteins were first discovered in the bacterium Myxococcus xanthus,6,18-20which turns red in the presence of light, as a result of the synthesis of carotenoids, but is pale yellow in the dark. However, carotenoids have also been found in a variety of other bacteria, including Streptomyces coelicolor21-23, Thermus thermophilus24-26and Bacillus megaterium27. Homologous sequences to the photoreceptor protein are found in the genomes of many bacteria and these proteins are probably widely distributed in non-photosynthetic bacteria.6,17,19222526 The photoreceptor proteins were named CarH in M. xanthus20, TtCarH25 or LitR24 in T. thermophilus and LitR27 in B. megaterium, but, for simplicity, and because the photoreceptor proteins are all very similar, in this review I shall use the designation CarH for the protein and carH for the gene.
The non-photosynthetic bacteria in which CarH has been identified vary widely in their habitat and metabolism. M. xanthus is a rod-shaped Gram-negative predatory bacterium. It is unable to synthesise B12 but can obtain B12 from its food and convert it to AdoCbl, presumably using an ATP:corrinoid adenosyltransferase enzyme because it has a gene encoding this enzyme.6T. thermophilus is a Gram-negative, extremely thermophilic bacterium (extremophile).28,29S. coelicolor is a Gram-positive filamentous bacterium.21,23B. megaterium is a Grampositive endospore-forming soil bacterium.27T. thermophilus28,29, S. coelicolor21,23and B. megaterium30are all capable of synthesising cobalamins de novo. M. xanthus and T. thermophilus are the bacteria that have been studied most intensively. In M. xanthus, the carH gene is found on the chromosome19 but in T. thermophilus it is found, together with genes for the later stages of B12 biosynthesis, on the megaplasmid, which is a large extrachromosomal element.24
The monomer of the photoreceptor protein CarH was found, by analysis of its primary structure, to consist of two regions joined by a short flexible linker chain: a C-terminal (carboxy) region, which can bind to cobalamins, and an N-terminal (amino) region, which can bind to DNA. The C-terminal end detects light and the N-terminal end allows gene expression for the production of carotenoids. The amino acid sequence at the N-terminal end of CarH is very similar to that of the DNA-binding domain of the N-termini of a family of transcriptional activators, known as the MerR-like proteins, which mediate responses to stress arising from exposure to toxic compounds or organic free radicals.20 The amino acid sequence of the carboxyl region of CarH is very similar to that of the MeCbl-binding domain of methionine synthase and shows a typical cobalamin-binding fingerprint sequence.6,13,18,19,31,32 However, the prosthetic group of CarH is AdoCbl, rather than MeCbl, as in methionine synthase.6 AdoCbl is bound to CarH through displacement of dbzm by the imidazole group of a conserved histidine (His193 in M. xanthus and His177 in T. thermophilus), similarly to the binding of MeCbl in methionine synthase.6
The active form of the CarH receptor protein is a tetramer containing four molecules of AdoCbl (Figure 2). CarH monomers oligomerise to form the tetramer only when AdoCbl is present. In the dark, the CarH tetramer binds to the DNA operator controlling carotenogenesis and prevents the action of RNA polymerase, thus blocking transcription of the proteins needed for synthesis of carotenes. In the presence of light at wavelengths at which AdoCbl absorbs light (360 nm, 438 nm or 540 nm), AdoCbl is photolysed, the tetramer dissociates into monomers and the gene repression is released. This process occurs only if the conserved histidine from the cobalamin-binding fingerprint sequence is present; mutants lacking this histidine, when histidine is replaced by alanine, do not show AdoCbl-dependent oligomerisation.6
It was later discovered that vitamin B12 also has a gene regulation function in the photosynthetic purple bacteria Rhodobacter capsulatus.33In the presence of light but the absence of oxygen, R. capsulatus produces large amounts of bacteriochlorophyll, and photosynthesis takes place through an anaerobic pathway. However, R. capsulatus can also grow aerobically in the dark.34 In the presence of sufficient oxygen, when the anaerobic pathway is not needed, the pathway is repressed because it also produces destructive singlet oxygen.17,33 The protein CrtJ represses the synthesis of bacteriochlorophyll and is linked to the protein AerR, which is also an aerobic repressor of photosynthesis genes.35 The AerR protein uses AdoCbl or MeCbl as its cofactor but does not bind to either.17,33 It was proposed (Figure 3) that AdoCbl or MeCbl is converted to hydroxocobalamin (HOCbl) in the presence of light, which is followed by binding of HOCbl to AerR, then binding of HOCbl/AerR to the gene repressor CrtJ, and finally release of the repression of the genes for synthesis of bacteriochlorophyll so that anaerobic photosynthesis can take place. AerR does not bind to AdoCbl or MeCbl but does bind to their photolysis product, HOCbl, and can therefore also be considered as a light-sensing protein.17,33
The AerR protein is similar to CarH in that it is homologous to CarH and methionine synthase, has a typical cobalamin binding fingerprint sequence and contains a histidine (His145) which binds to the lower position of B12, displacing dbzm. It is different in that it binds only HOCbl (rather than AdoCbl or MeCbl) and does not contain any DNA-binding region, but instead interacts with the protein CrtJ. AerR binds to HOCbl very strongly, much more strongly than expected from an ionic interaction between cobalamin and a single histidine on AerR. It was proposed that the upper position of the cobalamin is occupied by a second histidine (His10) from AerR and that this bond is stronger than that to the lower histidine.33
X-ray crystal structure of the CarH photoreceptor protein
In 2015, Jost, Drennan and coworkers1 determined the X-ray crystal structure of the photoreceptor CarH from T. Thermophilus in three different states: two dark states, in one of which CarH is free and in the other is attached to DNA and a state in which CarH has dissociated from DNA after exposure to visible light (Figure 4).
In the free dark state, CarH is a tetramer with one molecule of AdoCbl bound to each monomer.1 Each monomer has one domain that attaches to DNA at the N-terminal end and a second domain that binds the light-sensing AdoCbl molecule at the C-terminal end. The N-terminal domain consists of a DNA-recognition helix and a β-hairpin wing. The C-terminal domain consists of a four-helix bundle and a Rossman fold cobalamin-binding region. AdoCbl, in which the bond length of the Co-C bond is 2.0 Ä, similarly to that of free AdoCbl (2.030 Å)36, is located between the four-helix bundle and the Rossman fold1. The upper ligand, 5'-deoxyadenosyl, is orientated towards the four-helix bundle, close to a tryptophan residue, Trp131, and the lower ligand dbzm has been displaced by a histidine ligand (His177) from the Rossman fold. The C-terminal domain is rigid but the N-terminal domain is more flexible. The tetramer assembles itself from the monomers only if AdoCbl is present. The tetramer can be considered as a dimer of dimers, in which each dimer has its monomers in a head-to-tail orientation with respect to the AdoCbl-binding domain. The monomers within the dimer are held together by many hydrogen bonds and electrostatic interactions from a variety of side-chains and from the 5'-deoxyadenosyl ligand. The dimers are joined through their AdoCbl-binding domains in a staggered fashion to give the tetramer, involving Gly160 and Gly192 on each dimer. The C-terminal domains of the monomers form the centre of the tetramer with the N-terminal domains on the exterior.1
It is interesting that the primary structure of the AdoCbl-binding domain of CarH is similar to that of the MeCbl-binding domain of methionine synthase, rather than to any of the enzymes using AdoCbl as a prosthetic group, especially as the adenosyl group is much larger than the methyl group.1,6,13,18,19,31,32 However, the cobalamin binding pocket in CarH is bigger than that in methionine synthase because the four-helix bundle is situated 2.5 Å further away from the cobalamin and the leucine in methionine synthase is replaced by a smaller valine (Val138) residue. The pocket also provides more opportunities for hydrogen bonding by replacing a valine with glutamic acid (Glu141) and for polar interactions by replacing a valine with histidine (His142) and phenylalanine with tryptophan (Trp131).1
In the model for the AdoCbl-based regulatory switch (Figure 2), the CarH tetramer binds to its cognate DNA operator, a 30-bp (base pair) portion of the region of DNA between the gene encoding CarH and the gene encoding CrtB (the operator controlling carotenogenesis). In the crystal structure of the second dark state, the CarH tetramer is bound to a large piece (26 bp) of the cognate DNA operator.1 Three of the four DNA-binding domains of the CarH tetramer bind to DNA, with the fourth domain being disordered and not visible in the crystal structure. The three detectable DNA-binding domains all face in the same direction but otherwise the structure is similar to that of the free dark state. All three visible DNA-binding domains are important in binding CarH to DNA, forming a variety of hydrogen bonds and electrostatic interactions between the protein and the phosphate backbone of the DNA. In addition, the recognition helix of each DNA domain on CarH inserts into the DNA major groove and a histidine from the β-hairpin wing fits into the DNA minor groove. The recognition helix of the middle DNA domain covers the promoter -35 element for the gene corresponding to the σA-associated bacterial RNA polymerase, in which σA is the protein subunit necessary for initiation of RNA synthesis37, and thus blocks access of the RNA polymerase to the promoter. (The promoter -35 element is so called because it is 35 nucleotides upstream, or counting backwards, from the transcription start site of σA.38)
In contrast to the dark-state tetramer, the light-state CarH protein is monomeric and contains cobalamin, but has lost the 5'-deoxyadenosyl group.1 When comparing the light-state CarH monomer with the corresponding dark-state monomer, it can be seen that the Co-C bond of AdoCbl has been broken and the 5'-deoxyadenosyl group is absent. The helical bundle, including Trp131, has moved into the vacant space left by the 5'-deoxyadenosyl group and is positioned very much closer to cobalamin. Very importantly, a histidine, His132, on the helical bundle has shifted into a suitable position to coordinate with Co(III) on the cobalamin through its imidazole side-chain.
Mechanism of action of the CarH regulatory switch
The X-ray crystal structures of the three CarH proteins are consistent with the AdoCbl-based regulatory switch proposed in Figure 2, but give a much more detailed insight into its mechanism. In the dark state, the lower ligand (dbzm) of AdoCbl is displaced by the imidazole group of His177 and is thus bound through this histidine to CarH. In the His177 → Ala mutant, this step is blocked and the regulatory switch cannot function.1,6 In the dark, when carotenogenesis is not needed, the CarH tetramer binds to the DNA operator, preventing the transcription of carotenogenesis proteins. The X-ray crystal structures show that the DNA-binding domains of CarH are conveniently located on the outside of the tetramer, whereas the AdoCbl-binding domains are each buried in a deep pocket. In the presence of visible light, when carotenogenesis is of advantage to the bacteria, the Co-C bond of the bound AdoCbl is broken and the 5'-deoxyadenosyl moiety drifts away. The change in conformation on the breaking of the Co-C bond causes the movement of the helical bundle into the pocket, bringing His132 into position to bind to Co(III) through its imidazole side-chain. The change of conformation disrupts the head-to-tail dimer interaction and disassembles the tetramer, causing CarH to dissociate from the DNA promoter and relieving the blockage of access of RNA polymerase to the promoter so that transcription is allowed.1
In conjunction with X-ray crystallography, Jost et al.1,7 have used other techniques to answer questions about the mechanism of photolysis of CarH-bound AdoCbl. Two of these questions are:
1. What are the products of photolysis of CarH-bound AdoCbl and how exactly is the Co-C bond in CarH-bound AdoCbl broken?
2. How is the Co(III) in the light-state CarH protein bonded to the second histidine (His 132) on the protein?
Both questions have already generated controversy and will be considered further here.
CarH-bound AdoCbl: Products of photolysis and the Co-C bond
The UV-visible spectrum of the CarH free dark state is very similar to that of free AdoCbl and is completely consistent with the imidazole group of histidine being the lower ligand.1
After photolysis in the presence of oxygen the CarH-bound cobalamin is in the Co(III) oxidation state1,7 but in the absence of oxygen the cobalamin is in the Co(II) oxidation state, as shown by UV-visible spectroscopy and electron spin resonance spectroscopy.7 Exposure of the CarH-bound cobalamin in the Co(II) oxidation state to oxygen, after anaerobic photolysis, gives the same Co(III) product as does aerobic photolysis. However, the organic product of photolysis of CarH-bound AdoCbl was completely unexpected. Jost, Drennan and coworkers7 have shown unequivocally by liquid chromatography-mass spectrometry and nuclear magnetic resonance spectroscopy that the organic photolysis product of AdoCbl-bound CarH under both anaerobic and aerobic conditions is solely 4',5'-anhydroadenosine. The product 4',5'-anhydroadenosine had never been observed previously in the photolysis of AdoCbl (see above). However 4',5'-anhydroadenosine has been observed in the thermolysis of AdoCbl in glycerol39-41, which could provide a model for the photolysis of CarH-bound AdoCbl.
Garr and Finke39,41 found that, in the highly viscous solvent glycerol, thermolysis of AdoCbl at 110 °C gives rise to 5% 4',5'-anhydroadenosine (in addition to 5',8-cycloadenosine and 5'-deoxyadenosine), but in the less viscous solvent, ethylene glycol, no 4',5'-anhydroadenosine is seen. Thermolysis of adenosylcobinamide (the 5-coordinate analogue of AdoCbl in which the nucleotide base has been removed) in ethylene glycol gives 4',5'-anhydroadenosine as a major product (33%).40,41 For both AdoCbl and adenosylcobinamide, the corrinoid partner of 4',5'-anhydroadenosine is Co(II).39-41 No 4',5'-anhydroadenosine is seen in thermolysis of AdoCbl42 or adenosylcobinamide43 in aqueous solution. A viscous solvent, which can act as a strong cage for Co(II) and the 5'-deoxadenosyl radical, appears to be necessary for the comparatively slow formation of 4',5'-anhydroadenosine. Garr and Finke39-41 proposed that, while in close proximity in the cage, the Co(II) and the 5'-deoxyadenosyl radical (generated by homolytic fission) undergo a β-elimination reaction to give hydridocobalamin and 4',5'-anhydroadenosine. Hydridocobalamin then rapidly decomposes to Co(II) and hydrogen (Scheme 2).39-41,44
Based on the results of Garr and Finke39-41, Jost and Drennan and coworkers7 have proposed the following CarH photolysis mechanisms (Scheme 3, Paths 1 and 2). In Path 1 of Scheme 3, CarH-bound AdoCbl first undergoes photolysis to give Co(II) and the 5'-deoxyadenosyl radical by homolytic fission of the Co-C bond. Then, while in close proximity in the extra strong cage provided by the protein, β-elimination between Co(II) and the 5'-deoxyadenosyl radical takes place to give the Co(III) hydride (still bound to the protein) and 4',5'-anhydroadenosine. Clearly, the CarH protein would provide a much stronger cage than a viscous solvent, which would account for 4',5'-anhydroadenosine being the sole product of photolysis of CarH-bound AdoCbl.7 There is a large amount of evidence in favour of Path 1 and this path is favoured by Jost and Drennan and colleagues7. Cage effects by proteins have been previously observed in the AdoCbl-dependent glutamate mutase45,46 and in the MeCbl-binding domain of methionine synthase47. In addition to thermolysis of AdoCbl and adenosylcobinamide39-41, homolytic fission followed by radical-mediated β-elimination is well documented in alkylcobalamins and alkylcobinamides48-50. The generation of 4',5'-anhydroadenosine (rather than more reactive species as in photolysis in vitro) as the organic partner of CarH photolysis may represent a safety mechanism in ensuring that the reactive 5'-deoxyadenosyl radical is not released.1,7
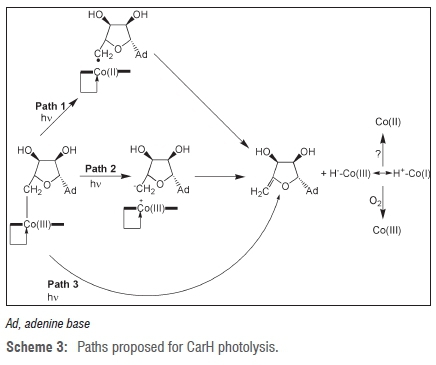
The intermediate hydridocobalamin, formulated
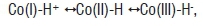
is an unstable species that rapidly dispropor tionates to give Co(II) and molecular hydrogen under anaerobic conditions44 and is presumably oxidised to Co(III) under aerobic conditions. Hydridocobalamin is difficult to characterise because it is extremely unstable. However, from cyclic voltammetry51, hydridocobalamin has pKa≈1 and is protonated on the 5,6-dimethybenzimidazole base as well as on the Co51-53. Thus, hydri-docobalamin can be considered as protonated Co(I) and can act as a Brönsted acid to protonate water:
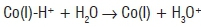
or possibly an amino acid side-chain on the protein.
Some doubt has been expressed about the structure of hydridocobalamin because 'hydridocobaloxime', which was proposed to have the same structure as hydridocobalamin54, has been found to be a dimeric Co(II) compound with a long Co-Co bond55. Cobaloximes are not good models for cobalamins in this case because they are relatively flat and can dimerise more readily than cobalamins. Cobalamins are prevented from dimerising in the same way by steric hindrance from their side-chains preventing a close enough approach of the two Co atoms.
Path 2 in the scheme of Jost et al.7 involves initial heterolytic cleavage of the Co-C bond to give Co(III) and the 5'-deoxyadenosyl anion, followed by β-elimination to give 4',5'-anhydroadenosine and hydridocobalamin. Kutta et al.56 propose a variant of Path 2 in which heterolysis of the Co-C bond gives the 5'-deoxyadenosyl anion and either hydridocobalamin or a five-coordinate positively charged Co(III) as intermediates on the route to 4',5'-anhydroadenosine and cob(II)alamin. It would seem that Path 2 would be highly unlikely. The 5'-deoxyadenosyl anion species appears to be completely unprecedented; a search of SciFinder produced no references compared with 284 for the 5'-deoxyadenosyl radical.57 An anion such as the 5'-deoxyadenosyl anion would be expected to be extremely unstable and would immediately rearrange or decompose to give a more stable species. This expectation has been observed in at least one case. A model reaction for heterolytic cleavage of the Co-C bond in CarH-bound AdoCbl is provided by the thermolysis of AdoCbl in aqueous solution at pH 7.0 and 85 °C. This reaction proceeded by 10% heterolysis, in which the organic products of heterolysis were only adenine and 2,3-dihydroxypentenal, which are decomposition products of 5'-deoxyadenosyl.42 The presence of exclusively 4',5'-anhydroadenosine (and no adenine) as the organic product of CarH photolysis strongly suggests that Path 2 is not operative in CarH photolysis.
Yet another path (Path 3) - concerted β-elimination by migration of a hydride ion (H-) to give hydridocobalamin and 4',5'-anhydroadenosine directly (that is a reaction not proceeding through a radical pair intermediate) - is theoretically possible. This type of reaction seems unlikely because it generally requires a vacant coordination site on the metal58 and has no precedent in organocobalamins.
How is Co(III) in the light-state CarH protein bonded to His 132?
After photolysis has taken place, the cobalamin moiety binds very strongly to light-state CarH, with the wild-type CarH forming a very stable complex with the cobalamin.1 The cobalamin-dependent light-sensing protein AerR in Rhodobacter capsulatus similarly forms a very stable complex with cobalamin and it has been proposed that this complex contains cobalamin bound to two histidines from the protein at both the top and bottom positions.17,33 However, this type of coordination of histidine is controversial because it has not been observed for free cobalamin (Marques et al.59 and personal observations). It also means that it is not possible to directly compare the UV-visible spectrum of the light-state CarH protein with that of free bis-histidine cobalamin in aqueous solution. The equilibrium constant for coordination to Co(III) of the first histidine (logK1 4.30) is favourable but coordination of the second histidine is much more difficult (logK2 <-1),59 so that the spectrum of the bis-histidine complex is not observed.59 However, the UV-visible spectra of cobalamin with histidine or imidazole in the upper position and dbzm in the lower position are almost indistinguishable,59 and the UV-visible spectrum of (mainly) the bis-imidazole complex of cobalamin can be observed as log K1=4.59 and log K2=0.6.59 The UV-visible spectrum of the light-state CarH protein is very similar to the UV-visible spectrum of the Co(III) cobalamin complex with two imidazole ligands binding through their N atoms in the upper and lower positions1,59 and is consistent with the substitution of cobalamin by two histidines from the protein1. Presumably, the cobalamin is constrained by the protein so that substitution of a second histidine becomes possible. The extreme stability of light-state CarH1 and AerR bound to cobalamin33, both of which show a cobalamin adduct in mass spectrometry1,33, suggests that the top histidine has a covalent bond to Co(III), rather than the ionic bond in the lower position1,33. It is possible that the top histidine is bound in the anion form, because imidazole bound as the anion has a larger logK than that for the neutral form (logK 4.60 rather than logK 4.30),59 which might account for the great stability of the light-state CarH protein. If His132 in the light-state CarH protein is mutated to alanine so that only one histidine can bind, the UV-visible spectrum of the corresponding lightstate CarH protein is very similar to the UV-visible spectrum of Co(III) cobalamin with only one imidazole1,59 or histidine59 ligand and it is less stable than the wild-type light CarH protein1.
Photochemistry of CarH
In an attempt to determine a more detailed photochemical mechanism for CarH, Kutta et al.56 performed photoexcitation experiments on the AdoCbl-bound CarH tetramer (ground-state CarH) and used transient UV-visible absorption spectroscopy on a femtosecond (10-15 s) to second timescale to follow the intermediates on the way to the light-state CarH product. They observed, in addition to Co(II) (and the presumed 5'-deoxyadenosyl radical), transient absorption spectra of at least eight intermediates and assigned these to possible structures, based on a model of the reaction pathway from ground-state CarH to light-state CarH. In their model, Intermediate A is the initial photoexcited state which, in their major pathway, immediately decays back to ground-state CarH either directly or through the Co(II)/5'-deoxyadenosyl radical pair, B. In their major pathway, the formation of the Co(II)/5'-deoxyadenosyl radical pair is unproductive, leading only to recombination to give AdoCbl. In their minor pathway, A converts to C, which has a spectrum that can be interpreted as a metal-to-ligand charge transfer complex, consisting of Co(III) with a partial positive charge and an incipient 5'-deoxyadenosyl anion. C then converts to D* (in the text) or C* (in their Figure 6),56 which is interpreted as a five-coordinate, positively charged Co(III) together with a 5'-deoxyadenosyl anion, then to D, in which the 5'-deoxyadenosyl anion has moved away from Co and has been replaced by His132, and lastly to E, which is similar to light-state CarH.56 Alternatively, they propose that Intermediate C corresponds to hydridocobalamin.56 The UV-visible spectrum of C is very similar in shape to that proposed for base-off hydridocobalamin,52 but is red-shifted relative to this spectrum. It is possible that C is the base-on form of hydridocobalamin but it is dangerous to speculate in this way (see below).
Further investigation is needed to elucidate the photochemical mechanism of CarH. Firstly, transient intermediate UV-visible spectra (300-700 nm) mainly tell us about the cobalamin partner and say very little about the 5'-deoxyadenosyl partner, which absorbs below 300 nm. It would perhaps be useful to extend the investigation into the UV region. Secondly, the identification of transient cobalamin intermediates in a mechanistic pathway, based only on their UV-visible spectra, is very difficult, unless suitable spectra of known species are available for comparison. For example, it is stated56 that the spectrum of Intermediate C appears to be similar to the S1 state seen in the photolysis of free MeCbl601,61. The S1 state in MeCbl photolysis has now been characterised as a d/p → p* metal-to-ligand charge-transfer state61 (consistent with theoretical density functional theory calculations)62, but was originally considered as 'a cob(III)alamin with a very weak axial ligand'60. The corresponding theoretical calculations for AdoCbl have not been completed to date but steady progress is being made with such calculations.61
Evolutionary implications
The discovery of the AdoCbl-dependent CarH photoreceptor and of the cobalamin-dependent AerR photoreceptor has many interesting implications. Genes for photoreceptors similar to CarH are found in a large number (>120) and variety of species of bacteria,6 including bacteria that can biosynthesise cobalamins and those that acquire cobalamins from their food. Genes similar to that for AerR are found in several species of purple photosynthetic bacteria.33 The presence of these genes suggests that B12-dependent photoreceptors are widespread in both non-photosynthetic and photosynthetic bacteria. Together with adenosycobalamin-dependent riboswitches, CarH and AerR (in combination with its partner CrtJ) may represent an evolutionarily very old function for cobalamin: interaction with and control of nucleic acids. AdoCbl may have been co-opted by proteins only at a later date. AdoCbl, which is essential for B12 enzymes in humans (as well as other animals and bacteria), may have first evolved as a regulatory switch for DNA and RNA.
Acknowledgements
I thank Dr Peter R. Johnson for his careful reading of the manuscript and the School of Chemistry, University of the Witwatersrand for a position as Emeritus Researcher.
References
1. Jost M, Fernández-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY-T, Kang G, et al. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature. 2015;526:536-541. http://dx.doi.org/10.1038/nature14950 [ Links ]
2. Banerjee R, Ragsdale SW. The many faces of vitamin B12: Catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209-247. http://dx.doi.org/10.1146/annurev.biochem.72.121801.161828 [ Links ]
3. Kräutler B. Organometallic chemistry of B12 coenzymes. In: Sigel A, Sigel H, Sigel RKO, editors. Metal-carbon bonds in enzymes and cofactors. Metal ions in life sciences. Volume 6. Cambridge: Royal Society of Chemistry; 2009. p. 1-51. http://dx.doi.org/10.1039/9781847559333-00001 [ Links ]
4. Matthews RG. Cobalamin- and corrinoid-dependent enzymes. In Sigel A, Sigel H, Sigel RKO, editors. Metal-carbon bonds in enzymes and cofactors. Metal ions in life sciences. Volume 6. Cambridge: Royal Society of Chemistry; 2009. p. 53-114. http://dx.doi.org/10.1515/9783110436587-006 [ Links ]
5. Gruber K, Puffer B, Kräutler B. Vitamin B12-derivatives - Enzyme cofactors and ligands of proteins and nucleic acids. Chem Soc Rev. 2011;40:4346- 4363. http://dx.doi.org/10.1002/chin.201145267 [ Links ]
6. Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elías-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci USA. 2011;108:7565-7570. http://dx.doi.org/10.1073/pnas.1018972108 [ Links ]
7. Jost M, Simpson JH, Drennan CL. The transcription factor CarH safeguards use of adenosylcobalamin as a light sensor by altering the photolysis products. Biochemistry. 2015;54:3231-3234. http://dx.doi.org/10.1021/acs.biochem.5b00416 [ Links ]
8. Law PY Wood JM. The photolysis of 5'-deoxyadenosylcobalamin under anaerobic conditions. Biochim Biophys Acta. 1973;331:451-454. http://dx.doi.org/10.1016/0005-2787(73)90032-4 [ Links ]
9. Hogenkamp HPC, Ladd JN, Barker HA. The identification of a nucleoside derived from coenzyme B12. J Biol Chem. 1962;237:1950-1952. [ Links ]
10. Hogenkamp HPC. A cyclic nucleoside derived from coenzyme B12. J Biol Chem. 1963;238:477-180. [ Links ]
11. Schwartz PA, Frey PA. 5'-Peroxyadenosine and 5'-peroxyadenosylcobalamin as intermediates in the aerobic photolysis of adenosylcobalamin. Biochemistry. 2007;46:7284-7292. http://dx.doi.org/10.1021/bi700077v [ Links ]
12. Hodgkin DC, Kamper J, Mackay M, Pickworth J, Trueblood KN, White JG. Structure of vitamin B12. Nature. 1956;178:64-66. http://dx.doi.org/10.1038/178064a0 [ Links ]
13. Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669-1674. http://dx.doi.org/10.1126/science.7992050 [ Links ]
14. Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, et al. How coenzyme B12 radicals are generated: The crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure. 1996;4:339-350. http://dx.doi.org/10.1016/s0969-2126(96)00037-8 [ Links ]
15. Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143-150. http://dx.doi.org/10.1093/nar/gkh167 [ Links ]
16. Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043-1049. http://dx.doi.org/10.1016/s1074-5521(02)00224-7 [ Links ]
17. Klug G. Beyond catalysis: Vitamin B12 as a cofactor in gene regulation. Mol Microbiol. 2014;91:635-640. http://dx.doi.org/10.1111/mmi.12490 [ Links ]
18. Cervantes M, Murillo FJ. Role for vitamin B12 in light induction of gene expression in the bacterium Myxococcus xanthus. J Bacteriol. 2002;184: 2215-2224. http://dx.doi.org/10.1128/jb.184.8.2215-2224.2002 [ Links ]
19. Elías-Arnanz M, Fontes M, Padmanabhan S. Carotenogenesis in Myxococcus xanthus: A complex regulatory network. In: Whitworth DE, editor. Myxobacteria: Multicellularity and differentiation. Washington, DC: ASM Press; 2008. p. 211-225. http://dx.doi.org/10.1128/9781555815677.ch12 [ Links ]
20. Pérez-Marín MC, Padmanabhan S, Polanco MC, Murillo FJ, Elías-Arnanz M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol Microbiol. 2008;67:804-819. http://dx.doi.org/10.1111/j.1365-2958.2007.06086.x [ Links ]
21. Takano H, Obitsu S, Beppu T, Ueda K. Light-induced carotenogenesis in Streptomyces coelicolor A3(2): Identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J Bacteriol. 2005;187:1825-1832. http://dx.doi.org/10.1128/jb.187.5.1825-1832.2005 [ Links ]
22. Takano H, Beppu T, Ueda K. The CarA/LitR-family transcriptional regulator: Its possible role as a photosensor and wide distribution in non-phototrophic bacteria. Biosci Biotechnol Biochem. 2006;70:2320-2324. http://dx.doi.org/10.1271/bbb.60230 [ Links ]
23. Takano H, Hagiwara K, Ueda K. Fundamental role of cobalamin biosynthesis in the developmental growth of Streptomyces coelicolor A3 (2). Appl Microbiol Biotech. 2015;99:2329-2337. http://dx.doi.org/10.1007/s00253-014-6325-z [ Links ]
24. Takano H, Kondo M, Usui N, Usui T, Ohzeki H, Yamazaki R, et al. Involvement of CarA/LitR and CRP/FNR family transcriptional regulators in light-induced carotenoid production in Thermus thermophilus. J Bacteriol. 2011;193:2451-2459. http://dx.doi.org/10.1128/jb.01125-10 [ Links ]
25. Díez AI, Ortiz-Guerrero JM, Ortega A, Elías-Arnanz M, Padmanabhan S, De la Torre JG. Analytical ultracentrifugation studies of oligomerization and DNA-binding of TtCarH, a Thermus thermophilus coenzyme B12-based photosensory regulator. Eur Biophys J. 2013;42:463-176. http://dx.doi.org/10.1007/s00249-013-0897-x [ Links ]
26. Takano H, Agari Y Hagiwara K, Watanabe R, Yamazaki R, Beppu T, et al. LdrP, a cAMP receptor protein/FNR family transcriptional regulator, serves as a positive regulator for the light-inducible gene cluster in the megaplasmid of Thermus thermophilus. Microbiology. 2014;160:2650-2660. http://dx.doi.org/10.1099/mic.0.082263-0 [ Links ]
27. Takano H, Mise K, Hagiwara K, Hirata N, Watanabe S, Toriyabe M, et al. Role and function of LitR, an adenosyl B12-bound light-sensitive regulator of Bacillus megaterium QM B1151, in regulation of carotenoid production. J Bacteriol. 2015;197:2301-2315. http://dx.doi.org/10.1128/jb.02528-14 [ Links ]
28. Lioliou EE, Pantazaki AA, Kiriakidis DA. Thermus thermophilus genome analysis; benefits and implications. Microbial Cell Factories. 2004;3:5-7. http://dx.doi.org/10.1186/1475-2859-3-5 [ Links ]
29. Henne A, Brüggeman H, Raasch C, Wiezer A, Hartsch T, Liesegang H, et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol. 2004;22:547-553. http://dx.doi.org/10.1038/nbt956 [ Links ]
30. Brey RM, Banner CDB, Wolf JB. Cloning of multiple genes involved with cobalamin (vitamin B12) biosynthesis in Bacillus megaterium. J Bacteriol. 1986;167:623-630. [ Links ]
31. Ludwig ML, Drennan CL, Matthews RT. The reactivity of B12 cofactors: The proteins make a difference. Structure. 1996;4:505-512. http://dx.doi.org/10.1016/s0969-2126(96)00056-1 [ Links ]
32. Ludwig ML, Matthews RT. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem. 1997;66:269-313. http://dx.doi.org/10.1146/annurev.biochem.66.1.269 [ Links ]
33. Cheng Z, Li K, Hammad LA, Karty JA, Bauer CE. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol Microbiol. 2014;91:649-664. http://dx.doi.org/10.1111/mmi.12491 [ Links ]
34. Tichi MA, Tabita FR. Interactive control of Rhodobacter capsulatus redox-balancing systems during phototrophic metabolism. J Bacteriol. 2001;183:6344-6354. http://dx.doi.org/10.1128/jb.183.21.6344-6354.2001 [ Links ]
35. Bauer C. Regulation of photosystem synthesis in Rhodobacter capsulatus. Photosynth Res. 2004;80:353-360. http://dx.doi.org/10.1023/b:pres.0000030440.99968.68 [ Links ]
36. Randaccio L, Geremia G, Nardin G, Würges J. X-ray structural chemistry of cobalamins. Coord Chem Rev. 2006;250:1332-1350. http://dx.doi.org/10.1016/j.ccr.2005.12.005 [ Links ]
37. Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441-146. http://dx.doi.org/10.1146/annurev.micro.57.030502.090913 [ Links ]
38. Wikipedia contributors. Promoter (genetics) [Wikipedia page]. No date [updated 2016 Feb 07; cited 2016 Mar 02]. Available from: https://en.wikipedia.org/w/index.php?title=Promoter_(genetics)&oldid=703782251 [ Links ]
39. Garr CD, Finke RG. Adocobalamin (AdoCbl or coenzyme B12) Co-C bond homolysis radical-cage effects: Product, kinetic, mechanistic, and cage efficiency factor (Fc) studies, plus the possibility that coenzyme B12-dependent enzymes function as "ultimate radical cages" and "ultimate radical traps". Inorg Chem. 1993;32:4414-4421. http://dx.doi.org/10.1021/ic00072a042 [ Links ]
40. Garr CD, Finke RG. Radical cage effects in adocobinamide (axial-base-off coenzyme B12): A simple method for trapping [Ado'Co"] radical pairs, a new β-H elimination product from the radical pair and measurement of an unprecedentedly large cage-recombination efficiency factor Fc> 0.94. J Am Chem Soc. 1992;114:10440-10445. http://dx.doi.org/10.1021/ja00052a045 [ Links ]
41. Finke RG. Coenzyme B12-based chemical precedent for Co-C bond homolysis and other key elementary steps. In: Kräutler B, Arigoni D, Golding BT, editors. Vitamin B12 and B12-proteins. Weinheim: Wiley-VCH; 1998. p. 383-102. http://dx.doi.org/10.1002/9783527612192.ch25 [ Links ]
42. Hay BP Finke RG. Thermolysis of the Co-C bond of adenosylcobalamin. 2: Products, kinetics, and Co-C bond dissociation energy in aqueous solution. J Am Chem Soc. 1986;108:4820-4829. http://dx.doi.org/10.1021/ja00276a020 [ Links ]
43. Hay BP, Finke RG. Thermolysis of the Co-C bond in adenosylcorrins. 3: Quantification of the axial base effect in adenosylcobalamin by the synthesis and thermolysis of axial base-free adenosylcobinamide. Insights into the energetics of enzyme-assisted cobalt-carbon bond homolysis. J Am Chem Soc. 1987;109:8012-8018. http://dx.doi.org/10.1021/ja00260a011 [ Links ]
44. Schrauzer GN, Holland RJ. Hydridocobalamin and a new synthesis of organocobalt derivatives of vitamin B12. J Am Chem Soc. 1971;93:4060-4062. http://dx.doi.org/10.1021/ja00745a048 [ Links ]
45. Sension RJ, Cole AG, Harris AD, Fox CC, Woodbury NW, Lin S, et al. Photolysis and recombination of adenosylcobalamin bound to glutamate mutase. J Am Chem Soc. 2004;126:1598-1599. http://dx.doi.org/10.1021/ja0396910 [ Links ]
46. Sension RJ, Harris AD, Stickrath A, Cole AG, Fox CC, Marsh ENG. Time-resolved measurements of the photolysis and recombination of adenosylcobalamin bound to glutamate mutase. J Phys Chem B. 2005;109:18146-18152. http://dx.doi.org/10.1021/jp052492d [ Links ]
47. Jarrett JT, Drennan CL, Amaratunga M, Scholten JD, Ludwig ML, Matthews RG. A protein radical cage slows photolysis of methylcobalamin in methionine synthase from Escherichia coli. Bioorg Med Chem. 1996;4:1237-1246. http://dx.doi.org/10.1016/0968-0896(96)00119-8 [ Links ]
48. Baldwin DA, Betterton EA, Chemaly SM, Pratt JM. The chemistry of vitamin B12. Part 25. Mechanism of the β-elimination of olefins from alkylcorrinoids; evidence for an initial homolytic fission of the Co-C bond. J Chem Soc Dalton Trans. 1985:1613-1618. http://dx.doi.org/10.1039/dt9850001613 [ Links ]
49. Kim S-H, Chen HL, Feilchenfeld N, Halpern J. Thermal decomposition and cobalt-carbon bond dissociation energies of organocobalamins: Neopentyl-, (cyclopentylmethyl)-, (cyclohexylmethyl)-, (tetrahydrofurfuryl)- and ((tetrahydro-2H-pyryl)methyl)cobalamin. J Am Chem Soc. 1988;110:3120-3126. http://dx.doi.org/10.1021/ja00218a021 [ Links ]
50. Pratt JM. Coordination chemistry of the B12 dependent isomerase reactions. In: Dolphin D, editor. B12. Volume 1. New York: Wiley; 1982. p. 326-392. [ Links ]
51. Lexa D, Savéant J-M. Brönsted basicity of vitamin B12s. J Chem Soc Chem Comm. 1975; 872-4. http://dx.doi.org/10.1039/c39750000872 [ Links ]
52. Chemaly SM, Pratt JM, The chemistry of vitamin B12. Part 24. Evidence for hydride complexes of cobalt(III) corrinoids. J Chem Soc Dalton Trans. 1984:595-599. http://dx.doi.org/10.1039/dt9840000595 [ Links ]
53. Pratt JM. The roles of Co, corrin and protein. I. Co-ligand bonding and the trans effect. In: Banerjee R, editor. Chemistry and biochemistry of B12. New York: Wiley; 1999. p. 73-112. [ Links ]
54. Schrauzer GN, Holland RJ. Hydridocobaloximes. J Am Chem Soc. 1971;93:1505-1506. http://dx.doi.org/10.1021/ja00735a040 [ Links ]
55. Lacy DC, Roberts GM, Peters JC. The cobalt hydride that never was: Revisiting Schrauzer's "hydridocobaloxime". J Am Chem Soc. 2015;137:4860-4864. http://dx.doi.org/10.1021/jacs.5b01838 [ Links ]
56. Kutta RJ, Hardman SJO, Johannissen LO, Bellina B, Messiha HL, Ortiz-Guerrero, et al. The photochemical mechanism of a B12-dependent photoreceptor protein. Nat Commun. 2015;6:7907-7918. http://dx.doi.org/10.1038/ncomms8907 [ Links ]
57. SciFinder. American Chemical Society (ACS) [homepage on the Internet]. No date [cited 2015 Nov 30]. Available from: https://scifinder.cas.org/scifinder/view/scifinder/scifinderExplore.jsf [ Links ]
58. Kochi JK. Organometallic mechanisms and catalysis. New York: Academic Press; 1978. p. 246. http://dx.doi.org/10.1016/b978-0-12-418250-9.50017-6 [ Links ]
59. Marques HM, Marsh JH, Mellor JR, Munro OQ. The coordination of imidazole and its derivatives by aquocobalamin. Inorg Chim Acta. 1990;170:259-269. http://dx.doi.org/10.1016/s0020-1693(00)80484-3 [ Links ]
60. Walker LA, Jarrett JT, Anderson NA, Pullen SH, Matthews RG, Sension RJ. Time-resolved spectroscopic studies of B12 coenzymes: The identification of a metastable cob(III)alamin photoproduct in the photolysis of methylcobalamin. J Am Chem Soc. 1998;120:3597-3603. http://dx.doi.org/10.1021/ja974024q [ Links ]
61. Rury AS, Wiley TE, Sension RJ. Energy cascades, excited state dynamics, and photochemistry in cob(III)alamins and ferric porphyrins. Acc Chem Res. 2015;48:860-867. http://dx.doi.org/10.1021/ar5004016 [ Links ]
62. Lodowski P Jaworska M, Andruniów T, Garabato BD, Koslowski PM. Mechanism of Co-C photolysis in the base-on form of methylcobalamin. J Phys Chem A. 2014;118:11718-11734. http://dx.doi.org/10.1021/jp508513p [ Links ]
 Correspondence:
Correspondence:
Susan Chemaly
susan.chemaly@wits.ac.za
Received: 06 Apr. 2016
Revised: 07 June 2016
Accepted: 30 June 2016














