Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.111 no.11-12 Pretoria Nov./Dez. 2015
http://dx.doi.org/10.17159/sajs.2015/20140233
RESEARCH ARTICLE
Incidence of non-typhoidal Salmonella in poultry products in the North West Province, South Africa
Roseline Y. Olobatoke; Sendros D. Mulugeta
Department of Animal Science, School of Agricultural Sciences, North West University, Mmabatho, South Africa
ABSTRACT
This study was conducted to evaluate the incidence of non-typhoidal Salmonella (NTS) serotypes in raw and ready-to-eat (RTE) broiler products in the North West Province of South Africa. A total of 120 raw broiler samples, 40 samples of polonies and 20 samples of smoked viennas were obtained from retail points in major cities and towns in the province. Samples were subjected to aerobic plate count and later screened for the presence of NTS using phenotypic and genotypic techniques. The average bacterial count in raw products was 3.1 x 105 cfu/g whereas bacterial contamination of RTE products was 1.8 x 103 cfu/g. The average recovery rate of NTS species from raw broiler products was 12.5% and the serotypes identified were S. typhimurium (46.4%), S. enteritidis (30.9%) and S. newport (22.9%). No NTS was recovered from the RTE products. However, S. typhimurium was the predominant serotype in whole carcasses whereas S. enteritidis and S. newport were prevalent in chicken parts. Out of the 160 presumptive NTS isolates screened by polymerase chain reaction (PCR), 140 (87.5%) were confirmed for the presence of the Salmonella-specific invA gene. In addition, 115 (82.4%) of the confirmed isolates harboured the plasmid spvC gene. Random Amplified Polymorphic DNA (RAPD) fingerprinting of isolates using RAPD 1 and RAPD 3 primers, revealed some inter- and intra-serotype genetic diversity among isolates, suggesting varying sources of contamination. The results of this study represent the first report on the incidence and prevalent serotypes of NTS in chicken products in the North West Province of South Africa.
Keywords: food-borne salmonellosis; broiler; invA; spvC; fingerprinting
Introduction
Non-typhoidal salmonellosis is an important public health problem worldwide and particularly in sub-Saharan Africa where it commonly manifests as gastroenteritis and/or bloodstream infections in both children and adults.1 The gastroenteritis form, which may sometimes be self-limiting, is commonly found in industrialised countries. Immunocompromised individuals including patients with HIV, cancer or diabetes, are at higher risk of non-typhoidal Salmonella (NTS) bacteraemia and often develop focal infections such as meningitis, septic arthritis, pneumonia and osteomyelitis.2 Although more than 2500 serovars of Salmonella enterica have been reported, S. typhimurium and S. enteritidis are identified as the commonest causes of human infection.3,4 A retrospective study in one hospital in the Democratic Republic of Congo from 2002 to 2006, revealed that NTS caused 59% of bacteraemia in children. Salmonella typhimurium and Salmonella enteritidis were responsible for 82% of the cases.5 In Mozambique, NTS were reported to account for 120 cases of childhood bacteraemia per 100 000 persons/year.6 Contaminated poultry meat and eggs, among other factors, have been implicated as vehicles of transmission for these hardy pathogens.7 For these reasons, there have been numerous studies focusing on assessing the incidence/prevalence of NTS strains in chicken carcasses and other meat products.7-9
South Africa has witnessed a tremendous increase in chicken meat consumption.10 Concurrently, the Enteric Disease Reference Unit of the National Institute for Communicable Diseases noted an increasing number of NTS isolates despite the fact that human salmonellosis cases are rarely reported. Evidence of these occurrences are the outbreaks of food-borne illnesses in Mpumalanga Province of South Africa incriminating NTS serotypes. One of the outbreaks involved the consumption of meals prepared with poultry products.11,12 These outbreaks indicate the presence of NTS in South Africa, which may be an issue of public health concern. A few investigations have been conducted in South Africa to ascertain the contamination of chicken carcasses and ready-to-eat foods from retail stores, with various pathogenic bacteria including Salmonella.13,14However, the incidence of NTS in broiler products in the North West Province (NWP) has not been established despite the fact that the NWP is one of the provinces with the largest production and distribution of broilers in the country. The objective of this study therefore was to screen raw and ready-to-eat broiler products obtained from the NWP of South Africa for incidence of NTS contamination.
Materials and methods
Experimental design
The study was cross-sectional with sampling based on two shop types and three product groups. The sampling, which lasted for 6 months, commenced in October 2010 and ended in March 2011, corresponding with the summer months in the study area.
Sample size
Primary population size to be sampled was determined according to the following formula15:

where p is the expected prevalence; L is the accepted error and Za is the value for normally distributed data at a confidence level α.
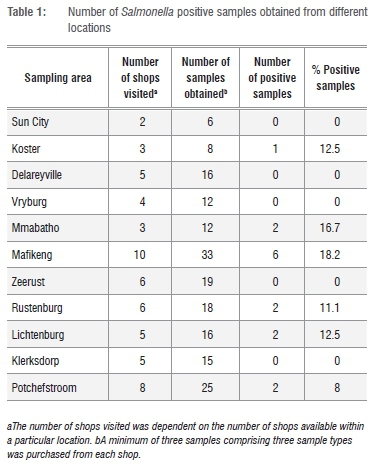
Based on an expected prevalence of 40% and a confidence level of 90%, a total of 57 shops were sampled, irrespective of the source of their products (i.e. either from within or outside the province). A total of 180 samples was obtained for analysis based on the purchase of a minimum of three product groups from each shop, and keeping 5% above the actual sample size to prevent the risk of sample loss during analysis (Table 1). Temperature of samples at point of purchase ranged from -20 °C to 4 °C. For all the procedures involved in the analysis, Salmonella typhimurium ATCC 14028 was used as a positive control strain whereas E. coli ATCC 25922 was used as a negative control. All culture, isolation and biochemical screening techniques were carried out using the MFHPB-20 procedure16 with appropriate modifications.
Determination of total bacterial count in samples
Briefly, 25 g of each sample was aseptically removed and homogenised with 225 mL of 2% buffered peptone water (BPW) in a stomacher bag (Nasco, Swedesboro, NJ, USA). Then 1 mL of the homogenate was transferred to Eppendorf tubes and serial dilutions were made using 2% BPW. An aliquot (0.1 mL) of each dilution was plated on plate count agar (Merck, SA) by spreading, and incubated at 37 °C for 24 h after which colonies were counted and recorded as cfu/g sample.
Isolation of Salmonella species from broiler carcasses and sausages
For broiler carcasses, each sample was aseptically removed from the package and transferred to a sterile plastic bag. Then 150 mL of 2% BPW was added to each bag and the mixture was shaken constantly for 2 min to obtain carcass rinse. Thereafter, 25 mL of the rinse liquid was transferred to a Whirl-pak bag (Nasco, USA) and another 75 mL of 2% BPW was added. Chicken sausages were aseptically homogenised with 225 mL of double strength (4%) BPW in a stomacher bag (Nasco, USA). Each bag was sealed and incubated at 37 °C for 18 h. Following incubation, contents of each Whirl-pak bag (Nasco, USA) was homogenised and 0.1 mL aliquot of the mixture was transferred to 10 mL of Rappaport-Vassiliadis broth (Merck, SA) for selective enrichment of Salmonella. The broth was incubated at 42 °C for 24 h and then kept at room temperature for 24 h.17 Thereafter, a loopful of each broth sample was streaked on xylose lysine desoxycholate (XLD) agar (Merck, SA) and Salmonella Shigella agar (Merck, SA) and incubated at 37 °C for 24-36 h. Suspected colonies were further purified on XLD agar and thereafter preserved on nutrient agar (Merck, SA) slants for further analysis.
Biochemical screening of presumptive isolates
All suspect colonies from the XLD agar culture were inoculated on triple sugar iron agar (Merck, SA) slant and incubated at 37 °C for 24 h. Presumptive Salmonella isolates from the triple sugar iron test were further confirmed using the API-20E test (bioMerieux Inc., SA) performed according to the manufacturer's instructions.
Serological identification
Isolates showing typical Salmonella biochemical reactions were sero-typed by the slide agglutination test using Salmonella specific polyvalent antisera (Davies Diagnostics, SA) for O and H antigens. The test was performed according to the manufacturer's instructions.
Molecular characterisation of isolates
DNA extraction
Genomic and plasmid DNA were extracted using the Zymo® kit (Inqaba Biotech, SA) and alkaline lysis method,18 respectively. The quantity and purity of all extracted DNA were estimated using a UV visible spectrophotometer (model S-22, Boeco, Germany) after which the integrity of the DNA was checked on standard submarine gel electrophoresis using 0.8% (w/v) agarose. The isolated DNA was stored at -20 °C until use.
PCR of extracted DNA
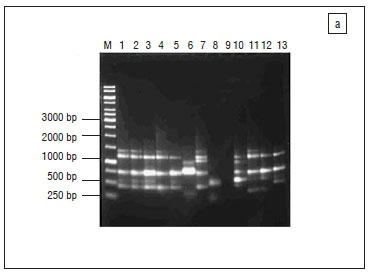
The 16S rRNA gene fragments of all isolates were amplified,19 after which presumptive Salmonella isolates were screened for the presence of invasion (invA) and virulence (spvC) genes for identity confirmation.20 All the oligonucleotide primers used for polymerase chain reactions (PCR) were obtained from Inqaba Biotech, South Africa, and details of the sequences and cycling conditions are shown in Table 2. All PCR reactions were prepared in 25 μL volumes consisting of 1 μg/μL of the template DNA, 50 pmol of each oligonucleotide primer set, 1x PCR master mix, 1U Taq DNA polymerase and nuclease free distilled water.
Amplifications were performed using a Peltier Thermal Cycler (model Dyad™ DNA Engine). A reaction blank containing all the components of the reaction mixture except the template DNA was included in each PCR procedure to check for contamination. Thereafter, 10μL of the PCR amplicons was analysed by standard submarine gel electrophoresis using 1% (w/v) agarose at 60 V for 6 h. Lambda DNA molecular weight marker was the gene ruler used in electrophoretic analyses. Electrophoresis was conducted in a horizontal Pharmacia biotech equipment system (model Hoefer HE 99X; Amersham Pharmacia Biotech, Sweden) using 1x TAE running buffer (40 mM Tris, 1 mM EDTA and 20 mM glacial acetic acid, pH 8.0). The gels were stained in ethidium bromide (0.1 mg/mL) for 10- 15 min and later visualised under UV light.18 A Gene Genius Bio imaging system (Syngene, Synoptics; UK) was used to capture the image using GeneSnap (version 6.00.22) software. Amplicons which showed clear bands for 16S rRNA and invA genes were purified and sequenced at Inqaba Biotech (South Africa), for strain identification of the isolates, using the ABI 3500 XL sequencer. The search for amino acid homology of the sequence results was done using the NCBI Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/).
RAPD-PCR
Confirmed Salmonella isolates identified through the expression of the Salmonella-specific invA gene were further subjected to fingerprinting by random amplified polymorphic DNA (RAPD)-PCR to evaluate the genetic diversity among the isolates. Oligonucleotides of short sequence were used21,22 and the PCR reactions were prepared as stated above. Primer details and PCR conditions are shown in Table 2. Aliquots (10 μL) of the resulting amplicons were characterised on 1.2% agarose gel electrophoresed at 60 V for 6 h. Isolates were later scored for the presence or absence (1 or 0) of each band on agarose gel.22
Data analysis
To obtain the cfu/g of sample for total bacterial count, colonies on plate count agar were counted and compared with the dilution factor.23 All Salmonella populations were transformed to base 10 logarithms before analysis. Recovery rate of Salmonella isolates was calculated using a previously described equation.24 Means in recovery rates between locations, shop and product types were compared using the Student's t-test of SPSS (SPSS 10.0 for windows, SPSS Chicago, IL, USA). Differences were considered significant at p<0.05.

For cluster analysis, the distance matrices, means and standard deviations were calculated using Statistica and the result was used to construct a phylogenetic relationship among isolates by the neighbour-joining method of Saitou and Nei25.
Results and discussion
Total bacterial count of samples
On average, total bacterial count of butchery samples (2.2 x 105 cfu/g) was higher than that of supermarket samples (1.4 x 104 cfu/g). Similarly, raw chicken parts were more contaminated (7.6 x 105 cfu/g) than whole chicken carcasses (2.5 x 105 cfu/g). RTE products were the least contaminated (1.8 x 103 cfu/g) perhaps because the products had been subjected to further processing. However, the level of microbial contamination of all analysed products was within the recommended range for some developed countries such as United Kingdom (104-105 cfu/g) and Australia (106 cfu/g).26 Sources of contamination of raw poultry products vary and could originate from the live birds, processing procedures or from the environment.27 The higher microbial contamination rate of butchery samples and raw chicken portions when compared with supermarket products might be an indication of poor hygienic conditions in the processing environment. This suggests a lack of strict hygiene control measures during product processing and could have a public health implication on the consumers as poultry are usually contaminated with a relatively high frequency of pathogenic bacteria. Indeed, raw poultry products have been reported in quite a number of human food poisoning cases, particularly following handling, undercooking or mishandling of the cooked products.28,29 Furthermore, high level of product contamination, such as recorded in some samples in the current study, could facilitate product spoilage, particularly when organisms such as Pseudomonas are involved.
Incidence of non-typhoidal Salmonella in broiler products
Out of 11 locations sampled, Salmonella was recovered from products obtained from 6 (54.5%) of the locations (Table 1). The difference in Salmonella isolation rates between positive locations was significant (p=0.009) with Mafikeng having the highest rate. Mafikeng is a major town with about the highest number of retail shops in the province. As a result, the number of shops visited and samples obtained from Mafikeng in this study were more than other locations and this could be responsible for the difference in Salmonella isolation rates between locations. Salmonella spp. was not recovered from RTE products whereas average recovery rate in raw broiler products was 12.5%. This rate is lower than the 19.5% reported in the Gauteng Province of South Africa13, 20% in the USA30, 30% in Canada31, 39.7% in Mexico32
and 60% in Portugal8. Other countries such as Zambia33, Saudi Arabia34, Turkey7 and Sudan9, however, recorded lower incidences of NTS (4.7%, 5.92%, 8% and 9.2%, respectively) in broiler products than noted in the current study. Incidence of Salmonella contamination was higher in whole chicken carcasses (30%) than chicken parts (20%). This was contrary to the observations of Chaiba et al.35 and Moussa et al.34 who noted higher incidences of NTS in chicken cuts in Morroco and Sudan respectively. The presence and distribution of NTS serovars vary from region to region depending on the sampling plan and detection limits of the methodology employed.36
Butchery products were more contaminated with NTS (32%) than supermarket products (18%). This result, though contrary to the report by Yang et al.37, was in agreement with the findings of Chaiba et al.35 and could be the result of better hygienic standards maintained in supermarkets than butcheries. Frozen products had a higher NTS recovery rate (10.4%) than fresh products (8.8%). This may suggest initial high contamination of the frozen products with NTS. Although previous studies identified storage temperature as an important risk factor for pathogen growth and survival, the result of the current study suggests that freezing temperature range was not sufficient to inactivate the pathogens. It has been documented that freezing may not be regarded as a means of destroying food borne microorganisms. This is because low freezing temperatures of about -20 °C are less harmful to the pathogens than the median temperature ranges.38
Molecular characterisation of non-typhoidal Salmonella isolates
A total of 160 presumptive Salmonella isolates obtained from analysed broiler products were screened for the presence of invA and spvC genes, following the positive amplification of the 16S rRNA fragment. The invA gene was detected in 140 (87.5%) of the 160 screened isolates. Sequencing of the 16S rRNA amplicons of the remaining 20 isolates showed that they belonged to three other coliform species namely Klebsiella, Escherichia and Serratia. These could be the contaminants that showed biochemical and serological reactions similar to Salmonella. The invA gene, located on the Salmonella pathogenicity island 1 (SPI 1), is highly conserved in Salmonella species and encodes a type III secretion system (TTSS) that exports proteins in response to bacterial contact with epithelial cells.39 The invA gene operon is essential for full virulence in Salmonella.40The high detection of invA gene in the current study may be an indication of the potential pathogenicity of the isolated Salmonella strains and could be a cause for public health concern. Torpdahl et al.40 and Khan et al.41 similarly detected invA gene in all Salmonella isolates recovered from poultry products and orange juice, respectively. Analysis of the sequenced isolates in the current study revealed three NTS serotypes, namely, S. typhimurium (46.4%; n=65), S. enteritidis (30.7%; n=43) and S. newport (22.9%; n=32). The sequences of the three serotypes were deposited in GenBank with accession numbers JX859913, KC683709 and JX859912, respectively. The rate of isolation of the different NTS serotypes was in agreement with the reports from previous research.35,36 All the identified serotypes were isolated from both supermarket and butchery samples as well as whole carcasses and chicken parts at different rates (Figure 1). This shows that the isolated Salmonella serotypes may be circulating within the environment and possible evidence is the fact that some of the products were contaminated with multiple NTS serotypes, which may also indicate varying sources of product contamination.
Out of the 140 isolates confirmed for the presence of the invA gene, 115 (82.4%) isolates were found to harbour the spvC genes representing 71.9% of all the screened isolates. The spvC gene in Salmonella spp. interacts with the host immune system and is responsible for an increased growth rate in host cells.41 The prevalence of the spvC gene fragment was higher in isolates from butchery samples (71.4%) than supermarket samples (25.9%) and isolates from chicken parts (57.1%) than whole broiler carcasses (42.9%). The detection rate of the spvC gene in the current study agreed with the findings of Bolton et al.42 and Khan et al.20 who detected the gene fragment in 97% and 88% of Salmonella typhimurium DT104 strains, respectively. However, Khan et al.41 did not detect spvC gene in any of the Salmonella serotypes subjected to multiplex PCR. The recovery rate of S. typhimurium in this study was notably higher than that of other serotypes (S. enteritidis and S. newport), as opposed to the report of Khan et al.41 who did not recover S. typhimurium isolates in the juice samples analysed. Thus the disparity in the detection of the spvC gene fragment could possibly be related to the prevalence of the gene in S. typhimurium serotype.
Analysis of RAPD-PCR
The profiles generated by the RAPD-PCR were composed of 1-5 bands ranging between 0.25 kb and 1.4 kb. Both RAPD 1 and RAPD 3 primers each generated four banding profiles for S. typhimurium, three for S. newport and two profiles for S. enteritidis (Figure 2a and 2b). One strain of S. newport (SN1), isolated from chicken parts obtained from the butchery, showed no bands with both RAPD 1 and RAPD 3 primers. Similarly, a strain of S. typhimurium (ST2) isolated from supermarket whole broiler carcass, showed three bands with RAPD 1 but no bands with RAPD 3 primer. However, both strains produced one band of about 0.45 kb as did most of the isolates with RAPD 2 primer although the primer had very poor discriminatory power and was therefore not included in the analysis. Cluster analysis of the RAPD 1 profiles revealed three different RAPD types for S. newport and S. enteritidis whereas S. typhimurium was classified into four types (Figure 3a). RAPD 3 primer on the other hand, grouped S. typhimurium into four, S. newport into three and S. enteritidis into two RAPD types (Figure 3b). Although a few isolates of the same serotype showed either similar or same banding patterns, some level of heterogeneity was generally noticed in the isolates, irrespective of the sources (i.e. geographical location, shop or product type). This may again indicate varying sources of product contamination (e.g. live birds, water, environment or product handler). Furthermore, changes in bacterial genome may occur in the same serovar through plasmid acquisition/genetic mutations without manifesting phenotype alterations.43 Interestingly, a particular strain of S. typhimurium, isolated from a Mafikeng product namely chicken drumstick, was closely clustered with the positive control strain by both RAPD 1 and RAPD 3 primers. This attests to the efficiency of the Salmonella detection method employed in the current study. In spite of the heterogeneity manifested by the isolates however, it may still be concluded that most of the isolates have the same clonal origin.
Conclusion
The results of this study represent the first report on the incidence and prevalent serotypes of NTS in chicken products in the NWP It is, however, worthy to note that the three NTS serotypes isolated in this study were also the same serotypes identified in 49 NTS cases reported from the NWP to GERM-SA in 2011.44 This indicates that raw chicken products may serve as major vehicles contributing to foodborne salmonellosis in the NWP. Furthermore, multiple contamination of one sample by different serotypes, and the genetic diversity among isolates of the same or different serotypes (as expressed by RAPD fingerprinting) suggest varying sources of product contamination. This is a cause for public health concern and calls for adequate monitoring and establishment of effective strategies to control contamination along the production/ supply chain, in order to safeguard public health. The fact that NTS was not recovered from polonies and viennas (RTE products) in the current study may indicate effectiveness of employed RTE processing techniques in eliminating Salmonella contaminants from raw products. This report should provide confidence and reassurance to both RTE producers and consumers.
Acknowledgement
We thank the management of the North-West University, South Africa, for providing the financial and material resources for this work.
Authors' contributions
R.O. was responsible for the project design and execution and for writing the manuscript. S.M. was the project leader and was responsible for the data analysis.
References
1. Morpeth SC, Ramadhani HO, Crump JA. Invasive non-typhi Salmonella disease in Africa. Clin Infec Dis. 2009;49:606-611. http://dx.doi.org/10.1086/603553 [ Links ]
2. Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263-269. http://dx.doi.org/10.1086/318457 [ Links ]
3. Humphrey TJ. Public-health aspects of Salmonella infections. In: Wray C, Wray A, editors. Salmonella in domestic animals. Wallingford, England: CABI Publishing; 2000. p. 245-263. http://dx.doi.org/10.1079/9780851992617.0245 [ Links ]
4. Herikstad H, Motarjemi Y Tauxe RV. Salmonella surveillance: A global survey of public health serotyping. Epidemiol Infect. 2002;129:1-8. http://dx.doi.org/10.1017/S0950268802006842 [ Links ]
5. Vandenberg O, Nyarukweba DZ, Ndeba PM, Hendriksen RS, Barzilay EJ, Schirvel C, et al. Microbiological and clinical features of Salmonella species isolated from bacteremic children in Eastern Democratic Republic of Congo. Pediatr Infect Dis J. 2010;29:504-510. http://dx.doi.org/10.1097/inf.0b013e3181cd615a [ Links ]
6. Sigauque B, Roca A, Mandomando I, Morais L, Quinto L, Sacarlal J, et al. Community-acquired bacteraemia among children admitted to a rural hospital in Mozambique. Pediatr Infect Dis J. 2009;28:108-113. http://dx.doi.org/10.1097/INF.0b013e318187a87d [ Links ]
7. Dogru AK, Ayaz ND, Gencay YE. Serotype identification and antimicrobial resistance profiles of Salmonella spp. isolated from chicken carcasses. Trop Anim Health Prod. 2010;42:893-897. http://dx.doi.org/10.1007/s11250-009-9504-7 [ Links ]
8. Antunes P Réu C, Sousa JC, Peixe L, Pestana N. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int J Food Microbiol. 2003;82:97-103. http://dx.doi.org/10.1016/S0168-1605(02)00251-9 [ Links ]
9. El Hussein AA, Mayha M, Elmadiena N, Elsaid SM, Mohammed AMS, Muckle CA, et al. Prevalence of Salmonella enterica subspecies enterica serovars in Khartoum State, Sudan. Res J Microbiol. 2010;5:966-973. http://dx.doi.org/10.3923/jm.2010.966.973 [ Links ]
10. Mons G. Economy and imports pose biggest threats to African poultry industry. World Poult. 2010;26:6-7. [ Links ]
11. Smith AM, Gouws A, Hoyland G, Sooka A, Keddy KH. Outbreaks of food-borne disease - A common occurrence but rarely reported. S Afr Med J. 2007;97:1272. [ Links ]
12. National Institute for Communicable Diseases (NICD). Salmonella Virchow foodborne illness outbreak. Communicable Diseases Communique. Johannesburg: NICD; 2010. p. 3. [ Links ]
13. Van Nierop W, Dusé AG, Marais E, Aithma N, Thothobolo N, Kassel M, et al. Contamination of chicken carcasses in Gauteng, South Africa by Salmonella, Listeria monocytogenes and Campylobacter. Int J Food Microbiol. 2005;99:1-6. http://dx.doi.org/10.1016/j.ijfoodmicro.2004.06.009 [ Links ]
14. Christison CA, Lindsay D, Von Holy A. Microbiological survey of ready-to-eat foods and associated preparation surfaces in retail delicatessens, Johannesburg, South Africa. Food Control. 2008;19:727-733. http://dx.doi.org/10.1016/j.foodcont.2007.07.004 [ Links ]
15. Magnani R. Sampling guide. Food and Nutrition Technical Assistance Project (FANTA) Washington DC: Academy for Educational Development; 1997. p. 8-22. [ Links ]
16. Reid A. Isolation and identification of Salmonella from food and environmental samples. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 2009. p. 1-14. Available from: http://doc.mbalib.com/view/d2b3d2bfed24ef386714adab0de8c57c.html [ Links ]
17. Waltman WD. Methods for the cultural isolation of Salmonella. In: Wray C, Wray A, editors. Salmonella in domestic animals. Wallingford: Cabi Publishing; 2000. p. 355-372. http://dx.doi.org/10.1079/9780851992617.0355 [ Links ]
18. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. 2nd ed. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 1989. [ Links ]
19. Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165-172. http://dx.doi.org/10.1007/BF02529967 [ Links ]
20. Khan MAH, Chowdhoury AH, Mosaddik MA, Shajahan M. Multidrug resistant gene (S) harboring on a 20 Kb plasmid in Salmonella typhi that causes typhoid-enteric fever. Pak J Biol Sci. 2000;3:911-914. http://dx.doi.org/10.3923/pjbs.2000.911.914 [ Links ]
21. Quintaes BR, Leal NC, Reis EM, Hofer E. Optimization of randomly amplified polymorphic DNA-polymerase chain reaction for molecular typing of Salmonella enterica serovar Typhi. Rev Soc Bras Med Trop. 2004;37:143-147. http://dx.doi.org/10.1590/S0037-86822004000200006 [ Links ]
22. Habtamu-Taddele M, Rathore R, Dhama K, Agarwal RK. Epidemiological characterization of Salmonella gallinarum isolates of poultry origin in India, employing two PCR based typing methods of RAPD-PCR and PCR-RFLP. Asian J Anim Vet Adv. 2011;6:1037-1051. http://dx.doi.org/10.3923/ajava.2011.1037.1051 [ Links ]
23. McLandsborough LA. Food microbiology laboratory. Boca Raton, FL: CRC Press; 2004. [ Links ]
24. Charimba G, Hugo CJ, Hugo A. The growth, survival and thermal inactivation of Escherichia coli O157:H7 in a traditional South African sausage. Meat Sci. 2010;85:89-95. http://dx.doi.org/10.1016/j.meatsci.2009.12.010 [ Links ]
25. Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-425. [ Links ]
26. Dahal N. Prevalence and antimicrobial resistance of Salmonella in imported chicken carcasses in Bhutan [MSc dissertation]. Chiang Mai/Berlin: Chiang Mai University/ Freie University; 2007. [ Links ]
27. Cohen N, Ennaji H, Bouchrif B, Hassar M, Karib H. Comparative study of microbiological quality of raw poultry meat at various seasons and for different slaughtering processes in Casablanca (Morocco). J Appl Poult Res. 2007;16:502-508. http://dx.doi.org/10.3382/japr.2006-00061 [ Links ]
28. Geornaras I, De Jesus A, Van Zyl E, Von Holy A. Microbiological survey of a South African poultry processing plant. J Basic Microbiol. 1995;35:73-82. http://dx.doi.org/10.1002/jobm.3620350204 [ Links ]
29. Mead GC. Microbiological quality of poultry meat: A review. Braz J Poult Sci. 2004;6:135-142. http://dx.doi.org/10.1590/S1516-635X2004000300001 [ Links ]
30. White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, et al. The isolation of antibiotic-resistant Salmonella from retail ground meats. New Engl J Med. 2001;345:1147-1154. http://dx.doi.org/10.1056/NEJMoa010315 [ Links ]
31. Bohaychuk VM, Gensler GE, King RK, Manninen KI, Sorensen O, Wu JT et al. Occurrence of pathogens in raw and ready-to-eat meat and poultry products collected from the retail marketplace in Edmonton, Alberta, Canada. J Food Protec. 2006;69:2176-2182. [ Links ]
32. Zaidi MB, McDermott PF, Fedorka-Cray P Leon V Canche C, Hubert SK, et al. Nontyphoidal Salmonella from human clinical cases, asymptomatic children, and raw retail meats in Yucatan, Mexico. Clin Infec Dis. 2006;42:21-28. http://dx.doi.org/10.1086/498508 [ Links ]
33. Hang'ombe BM, Sharma RN, Skjerve E, Tuchili LM. Occurrence of Salmonella enteritidis in pooled table eggs and market-ready chicken carcasses in Zambia. Avian Dis. 1999;43:597-599. http://dx.doi.org/10.2307/1592662 [ Links ]
34. Moussa IM, Gassem MA, Al-Doss AA, Mahmoud WAS, Abdel-Mawgood AL. Using molecular techniques for rapid detection of Salmonella serovars in frozen chicken and chicken products collected from Riyadh, Saudi Arabia. Afr J Biotechnol. 2010;9:612-619. [ Links ]
35. Chaiba A, Fouzia RF, Abdelkader C, Rachida SB, Mouloud Z. Occurrence of Salmonella in chicken carcasses and giblets in Meknès-Morocco. Pak J Nutr. 2008;7:231-233. http://dx.doi.org/10.3923/pjn.2008.231.233 [ Links ]
36. Dominguez C, Gomez I, Zumalacarregui J. Prevalence of Salmonella and Campylobacter in retail chicken meat in Spain. Int J Food Microbiol. 2002;72:165-168. http://dx.doi.org/10.1016/S0168-1605(01)00638-9 [ Links ]
37. Yang B, Xi M, Wang X, Cui S, Yue T, Hao H, et al. Prevalence of Salmonella on raw poultry at retail markets in China. J Food Prot. 2011;74:1724-1728. http://dx.doi.org/10.4315/0362-028X.JFP-11-215 [ Links ]
38. Jay JM, Loessner ML, Golden DA. Protection of foods with low temperature. In: Jay JM, editor. Modern food microbiology, food science text series. 7th ed. New York: Springer Science and Business Media Inc.; 2005. p. 395-409. [ Links ]
39. Galán JE, Ginocchio C, Costeas P Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: Homology of InvA to members of a new protein family. J Bacteriol. 1992;17:4338-4349. [ Links ]
40. Torpdahl M, Skov MN, Sandvang D, Baggesen DL. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J Microbiol Methods. 2005;63:173-184. http://dx.doi.org/10.1016/j.mimet.2005.03.006 [ Links ]
41. Khan AA, Melvin CD, Dagdag EB. Identification and molecular characterization of Salmonella spp. from unpasteurized orange juices and identification of new serotype Salmonella strain S. enterica serovar Tempe. Food Microbiol. 2007;24:539-543. http://dx.doi.org/10.1016/j.fm.2006.09.002 [ Links ]
42. Bolton LF, Kelley LC, Lee MD, Fedorka-Cray PJ, Maurer JJ. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene, which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348-1351. [ Links ]
43. Baggesen DL, Christensen J. Distribution of Salmonella enterica serotypes and phage types in Danish pig herds. In: Bech-Nielsen S, Nielsen JP, editors. Proceedings of the Second International Symposium on Epidemiology and Control of Salmonella in Pork; 1997 Aug 20-22; Copenhagen, Denmark. Copenhagen: Federation of Danish Pig Producers and Slaughterhouses; 1997. p. 107-109. [ Links ]
44. Crowther-Gibson P Govender N, Keddy K, Perovic O, Quan V Von Gottberg A. Non-typhoidal Salmonella enterica (NTS). In: Germs-SA annual report. Johannesburg: NICD; 2011. p. 12-14 [ Links ]
 Correspondence:
Correspondence:
Roseline Olobatoke
Centre for Applied Radiation Science and Technology, Nor th West University
Private Bag X2046, Mmabatho 2735
South Africa
Email: yemisirose205@yahoo.com
Received: 16 July 2014
Revised: 10 Oct. 2014
Accepted: 23 Feb. 2015