Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.111 n.7-8 Pretoria Jul./Aug. 2015
http://dx.doi.org/10.17159/SAJS.2015/20150074
RESEARCH ARTICLE
Allele frequencies of AVPR1A and MAOA in the Afrikaner population
J. Christoff Erasmus; Anton Klingenberg; Jaco M. Greeff
Department of Genetics, University of Pretoria, Pretoria, South Africa
ABSTRACT
The Afrikaner population was founded mainly by European immigrants that arrived in South Africa from 1652. However, female slaves from Asia and Africa and local KhoeSan women may have contributed as much as 7% to this population's genes. We quantified variation at two tandem repeats to see if this historical founder effect and/or admixture could be detected. The two loci were chosen because they are in the promoters of genes of neurotransmitters that are known to be correlated with social behaviour. Specifically, arginine vasopressin receptor 1A's (AVPR1A) RS3 locus has been shown to correlate with age of sexual onset and happiness in monogamous relationships while the tandem repeat in the promoter of the monoamine oxidase A (MAOA) gene correlates with reactive aggression. The Afrikaner population contained more AVPR1A RS3 alleles than other Caucasoid populations, potentially reflecting a history of admixture. Even though Afrikaners have one of the lowest recorded non-paternity rates in the world, the population did not differ at AVPR1A RS3 locus form other European populations, suggesting a non-genetic explanation, presumably religion, for the low non-paternity rate. By comparing population allele-frequency spectra it was found that different studies have confused AVPR1A RS3 alleles and we make some suggestions to rectify these mistakes in future studies. While MAOA allele frequencies differed between racial groups, the Afrikaner population showed no evidence of admixture. In fact, Afrikaners had more 4-repeat alleles than other populations of European origin, not fewer. The 4-repeat allele may have been selected for during colonisation.
Keywords: admixture; founder effect; monoamine oxidase; arginine vasopressin receptor 1a; non-paternity
Introduction
The Afrikaner population of South Africa derives from about the same proportion of German, French and Dutch immigrants that came to the Cape from 1652 to 1806.1 Because the number of immigrants was finite, the Afrikaner population is considered a textbook example of a founder effect2, with many genetic diseases in overabundance compared with European populations3-5. However, as many as 5000 European men settled at the Cape between 1657 and 1866.6 As most of the immigrants were men, they occasionally married non-European women6,7 who were either slaves from Africa and India (including Indonesia and East Asia) or local Khoe and San (KhoeSan) women6-9. This practice is reflected genetically by the presence of non-European alleles in the Afrikaner population.10
The Afrikaner population fought several local wars against the KhoeSan, Xhosa, Zulus and British. It could be argued that these aggressive encounters may have been frequent and severe enough to have left traces of selection on the population. One gene that may have played an important role in this regard is monoamine oxidase A (MAOA) which breaks down serotonin and dopamine and which has been linked to increased reactive aggression.11-14
Reactive aggression in humans may be affected by the alleles they carry at the variable number of tandem repeats (VNTR) in the promoter region of the MAOA gene, which is located on the X chromosome at Xp11.23.11-15 These VNTRs occur in 2, 3, 3.5, 4 or 5 repeats of 30 base pairs (bp).13,16 These repeats can be classified as either high or low activity alleles, with the 2, 3 and 5 repeats constituting the low activity (MAOA-L) alleles and the 3.5 and 4 repeats the high activity (MAOA-H) alleles.12 The MAOA-H allele has a 2-10 times more effective transcription than the MAOA-L alleles.13,16,17 Others11,12,18 have determined that carriers of MAOA-L react more aggressively in provocational circumstances than their MAOA-H counterparts. The MAOA-H carriers better tolerated maltreatment and were also less likely to develop antisocial traits.18
Recent studies revealed a very low non-paternity rate of less than 1% in the Afrikaner population19,20 (see Greeff and Erasmus21 for an exception to the rule). Strong religious convictions, as was the case for Afrikaners22, have been suggested as an important determinant of marital fidelity23. However, the low rate of non-paternity in Afrikaners may have a genetic component: two studies point to the potential importance of arginine vasopressin receptor 1a (AVPR1A) in this context. Prichard et al.24 have shown that age of first sexual encounter is correlated to repeat length of alleles of AVPR1A. Similarly, Walum et al.25 have shown that certain alleles of AVPR1A seem to predispose their carriers to a less fulfilling monogamous life. Given Afrikaners' low non-paternity rates, it is of interest to quantify AVPR1A for this population.
AVPR1A is located at 12q14-15 and there are three polymorphic repeat regions in its 5' flanking region.26 One of these, a complex(CT)4-TT-(CT)8-(GT)24 repeat known as RS3, is 3625 bp upstream from transcription initiation26 and has been linked to human social behaviour in a number of studies. Given the early lead from voles in which longer microsatellite length results in higher levels of transcription27, functional magnetic resonance imaging showed that carriers of longer repeats had significantly stronger activation of their amygdala upon an emotional test28. Similarly, longer AVPR1A RS3 alleles were found to be significantly more transcribed in post-mortem examination of the hippocampal area of humans.29 From the behavioural side, male individuals with two long alleles are significantly more likely to have sexual intercourse before the age of 15 than male individuals with a short/long genotype.24 Individuals with longer alleles are also more likely to be altruistic in the dictator game.29 Other studies have linked specific alleles with altruism in pre-schoolers30, happiness in monogamous relationships25, social behaviour and autism31-34, musicality35-37, creative dance38 and eating attitudes39.
Both of these genes, AVPR1A and MAOA, could have unusual frequencies in the Afrikaner population because of the founder effect and/or admixture and may have affected the population's average behaviour. The aim of this study was to characterise the frequencies of AVPR1A's RS3 microsatellite and the MAOA VNTR alleles in the Afrikaner population and to compare them to other populations. However, comparing allele frequencies across populations highlighted a problem with standardised allele calls at the RS3 locus.
Materials and methods
Sample collection
Ethical clearance was obtained from the Ethics Committee of the Faculty of Natural and Agricultural Sciences, University of Pretoria (no. EC11912-065). A total of 200 male volunteers from the Afrikaner population were confirmed not to be fourth-degree relatives through self-supplied ancestries and the majority were clustered into 23 groups that are very distantly related by paternal ancestry (at least 14 degrees). Note that this is no more than can be expected of random individuals.9 These men considered themselves Afrikaners and have typical Afrikaner surnames. All volunteers completed an informed consent form and signed an agreement which stated that they understood that their DNA was going to be used for analysis and that they donated it willingly. The study adhered to the principles of the Declaration of Helsinki. Saliva samples were collected from participants using the Oragene-DNA self-collection kit supplied by DNA Genotek (Kanata, Ontario, Canada) and genomic DNA was isolated according to the manufacturer's instructions.
Genotyping
For both loci, the polymerase chain reaction (PCR) set-up consisted of 50 ng DNA, 1X AmpliTaq® 360 Buffer (Applied Biosystems, Foster City, CA, USA), 20 μΜ dNTPs, 250 μΜ MgCl2, 0.4 μΜ forward and reverse primers, 2% AmpliTaq® 360 GC Enhancer and 1.25 units AmpliTaq® 360, in a final reaction volume of 10 μL, and the PCR reactions were run in the 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA). The basic PCR cycle was repeated 35 times and included an initial denaturation step of 5 min at 95 °C and a final elongation step of 5 min at 72 °C.
MAOA
The genomic DNA of the promoter region in the MAOA gene was PCR amplified for subjects with the primer MAOaPT1 5'-ACAGCCTGACCGTGGAGAAG-3' and MAOaPB15'-GAACGGACGCTCCATTCGGA-3'.16 The cycling conditions were as follows: 95 °C denaturation step for 1 min followed by primer annealing at 62 °C for 1 min and elongation at 72 °C for 1 min. The PCR products were separated on 3% agarose gels, with a 20-bp ladder (Promega, Madison, WI, USA). Six bands were excised from the gel and purified with the High Pure PCR purification kit (Roche Diagnostics, Germany) and were successfully sequenced with BigDye (Applied Biosystems, Foster City, CA, USA). Cycle sequencing products were purified with ethanol precipitation and ran on a 3130xl genetic analyser (Applied Biosystems, Foster City, CA, USA). These sequences were aligned to the reference sequence from the National Centre for Biotechnology Information (GenBank: M89636.1) with BioEdit version 7.2.240 and three unique sequences were deposited in the European Nucleotide Archive (accession numbers LN813020 - LN813022). Based on size differences, cases with 3, 4 and 5 repeats were selected to serve as size markers for the identification of the remaining samples on the gel.
AVPR1A
The RS3 microsatellite of AVPR1A was amplified with a labelled forward primer 5'-6-FAM-TCCTGTAGAGATGTAAGTGC-3' and the reverse 5'-GTTTCTTTCTGGAAGAGACTTAGATGG-3'.32 Cycling conditions wereas follows: denaturation at 94 °C for 1 min, primer annealing at 54.7 °C for 45 s and elongation at 72 °C for 1 min. Amplicons were run on an ABI 3500 XL genetic analyser (Applied Biosystems, Foster City, USA). The final allele lengths were scored with GeneMapper software version 4.1.1 (Applied Biosystems, Foster City, USA). Individuals homozygous for alleles 332, 334, 336, 338, 340, 342 and 346 were sequenced with the PCR primers to determine the actual number of CT and GT repeats and the sequences were deposited in the European Nucleotide Archive (accession numbers LN812321 to LN812340).
Comparable allele frequencies
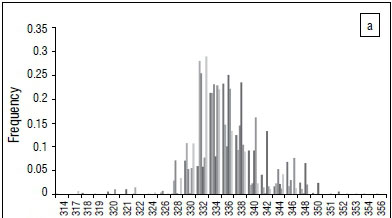
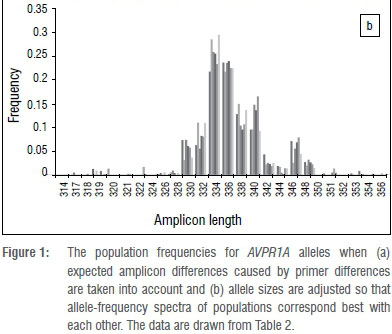
Like other microsatellites, AVPR1A RS3 allele calling can easily vary among studies because of different Taq, dye, polymers, size standards and machines, which prevents comparisons among studies. To complicate matters further, different studies have used primers that result in different sized amplicons. Working from the sequence published by Thibonnier et al.26 (AF208541), primer sets can result in an amplicon of 260 bp41, 324 bp25,28,32,37 (and the present study), 316 bp29-31,33,38,39 or 317 bp34, while others do not report primers used36. Fortunately, the allele spectrum for Caucasoids has a very characteristic profile that can be used to slide the alleles along so that they align well (compare Figure 1a and 1b). It can be seen that typically there are two alleles that are much more frequent than the others, and that there is another frequent allele that is 5 repeat units larger than the biggest common allele (Figure 1). We used this allele profile to make data sets comparable.
Statistical analysis
For both loci, pairwise FSTs were calculated between the populations using Arlequin version 3.5.1.2.42 For AVPR1A we also compared the Afrikaner's frequency of allele 334 to that of the other populations by first testing if the allele's frequency varied significantly over all five population samples (prop.test as implemented in R43). Then the two European populations32,37 and the two Israeli samples30,33 were combined and compared to each other and the Afrikaner population in a pairwise proportion test as implemented in R (pairwise.prop.test).43
Results
MAOA
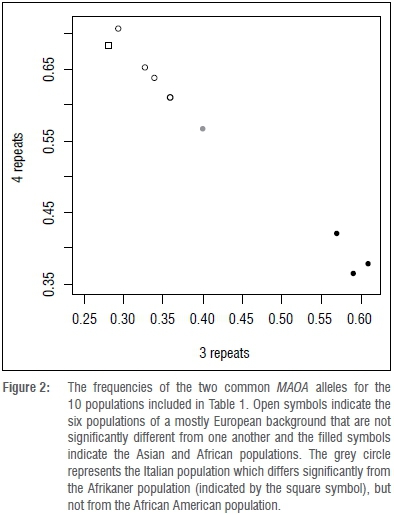
Sequencing confirmed that we were amplifying the correct DNA and we used these confirmed allele sizes as standards for electrophoresis. The frequencies of the observed alleles in the Afrikaner population and other populations are summarised in Table 1. The 3 and 4 repeat alleles were most frequent and varied across populations (Table 1; Figure 2). Pairwise FST values split the populations into two groups (Supplementary table 1 online). One group consisted of those of European descent for whom the 4-repeat allele was more common (Figure 2, clear symbols), and the other group consisted of those of African and Asian descent for whom the 3-repeat allele was more frequent (Figure 2, filled circles). Within each group, FST values were generally smaller than 0.01, and FST values between groups were mostly greater than 0.1 and significantly different (all p<0.001). A sample from Italy was significantly different from the Afrikaner population but not significantly different from the African American sample. Interestingly, two of the admixed populations - Hispanics and Afrikaners - did not fall in between European and non-European populations but had a higher frequency of the 4-repeat allele.

AVPR1A
Alleles that contained a combined number of 32 CT and GT repeats ran to a length of 334 on our machine (Table 2). We identified 20 alleles in the Afrikaner population whereas the other studies identified 15 or 16 alleles (Table 2). Afrikaners had a higher expected heterozygosity of 0.86 compared to values ranging from 0.83 to 0.84 (Supplementary table 2). For population comparisons it was impossible to align the allele frequencies of one Asian study34 with those of the Caucasoid populations because the allele frequency spectrum did not have the characteristic hallmarks of the locus in Caucasoid populations (Figure 1). As a result, no African or Asian populations could be compared to the Caucasoid populations. The frequencies of the adjusted allele sizes in other studies and ours are given in Table 2. With the allele sizes used as published and corrected for primer differences, pairwise FSTs were as high as 0.07 and all populations differed significantly from one another (Supplementary table 3). With the corrected sizes as given in Table 2, the FST values were all lower than 0.006 and mostly considerably lower (Supplementary table 4). The two Israeli samples30,33 were not significantly different from one another (p=0.775) but were significantly different from the other populations (all p<0.045), which in turn were not significantly different from one another (all p>0.11). The frequency of allele 334 differed among the population samples (χ2 = 16.723, df = 4, p=0.0022) as follows: Afrikaners = 0.21; Israeli 130 = 0.28; Israeli 233 = 0.29; British37 = 0.25; and American, mainly Caucasoid32 = 0.25. The pairwise test suggests that the Israeli sample had a significantly higher frequency of allele 334 than the Afrikaner (p=0.006) and European populations (p=0.010), but that the latter two did not differ significantly from each other (p=0.143).
Discussion
The two loci we considered provided two very different depictions. MAOA did not reveal any traces of admixture in the Afrikaner population as its allele frequency was displaced away from the African (as gauged from African American frequencies) and Asian populations rather than towards them (Figure 2). On the other hand, the AVPR1A showed an increased number of alleles in the Afrikaner population compared to other European populations, which could indicate the influence of admixture with older African and KhoeSan populations.6 Neither locus suggested a strong deviation from European frequencies caused by a founder effect.
The unexpected high frequency of the 4-repeat allele of MAOA in the Afrikaner population (Figure 2) requires an explanation. We need to take into account that the founder effect was more severe for female individuals in the population9; despite an influx of male individuals, there was no such influx of female individuals.6,9 In addition, because male individuals contribute only a single X chromosome, X chromosomes may have experienced a more severe bottleneck than other autosomal chromosomes. If we also consider that all non-European genetic contribution to this population was female derived,6,7,9 it seems questionable that the frequency could be skewed away from African and Asian frequencies. It also is interesting that one of the other populations with an admixed heritage, the Latinos from America, also had a higher frequency of the 4-repeat allele (Figure 2).
This finding may suggest that this allele may integrate more easily into a heterogeneous genetic background or that this small deviation simply stems from a small founder effect. Another explanation may be selection on the social phenotype of this allele. If MAOA-H (4-repeat allele) carriers are less likely to react aggressively in provocational circumstances12,13 and by extension be less likely to be berserkers in war scenarios, cope better with maltreatment18, and be less likely to develop antisocial behaviour11, then the allele may have been selected for in this founding population. It is, however, important to note that this allele's frequency is not significantly higher in the Afrikaner population and we should caution against over-interpreting this result as such.
For AVPR1A RS3, it is firstly important to make sure that alleles compared among studies are indeed the same. Because of the complex nature of AVPR1A RS, the various repeat alleles that run to the same length may differ in their proportions of GT and CT repeats.41 However, for a number of studies in which microsatellites in the promoter regions affect expression of the allele it seems to be the length rather than the content that is important.41 In addition to this complication, it is easy to systematically call alleles a number of base pairs shorter or longer when their identity is inferred from the rates of amplicon migration in different genetic analysers and/or when labelled with different dyes etc.; for this reason, it is important to standardise allele size in some way. We followed two approaches that were both effective. Comparable lengths can be obtained either by sequencing amplicons to confirm their length or by comparing allele-frequency spectra between populations. In this light, it is important to note that this study only considered Caucasoid populations, and the same characteristic peaks may not be observed in other populations. In fact, it was impossible to align the Asian study34 with the Caucasoid ones.
Confusion over which alleles are which is not trivial. The Israeli group has correctly linked their allele 327 to allele 334 from the Meyer-Lindenberg et al.28 and Kim et al.32 studies, but Walum et al.'s25 334 is in fact one base pair repeat longer than those in these other studies. As several researchers are comparing the effects of this locus on many behavioural patterns, it is important to have a gold standard to avoid confusion. We have compiled a ladder that can be used for such standardisation that is available on request. The confusion is not limited to comparisons of specific alleles; binning of alleles into groups of short, medium and long alleles can also lead to confusion. For instance, Prichard et al.24 classified short alleles as those that have 12-19 repeats, medium alleles as those with 20-21 repeats and long alleles as those with 22-29 repeats; however, these numbers of repeats are substantially lower than those observed in this and other26 studies (Table 2).
While the Afrikaner population had a lower frequency of the 334 allele, the frequency was not significantly lower and it would be premature to link their low levels of non-paternity with their low frequency of this allele. It is more likely that the Afrikaners' strong religious convictions24 could explain their low non-paternity rate19,20. The fact that the Israeli samples had a significantly higher frequency of allele 334 suggests that an investigation into marital happiness may be interesting for this population.
Acknowledgements
We thank the participants for providing samples, and two reviewers and the associate editor for their thought-provoking comments. This work was supported by a National Research Foundation (NRF) incentive grant 77256 to J.M.G. This work is based on research supported in part by a number of grants from the NRF (UID: 78566 (NRF RISP grant for the ABI3500)). The grantholders acknowledge that the opinions, findings and conclusions or recommendations expressed in any publication generated by NRF-supported research are those of the authors, and that the NRF accepts no liability whatsoever in this regard.
Authors' contributions
A.K. performed the work under the mentorship of J.C.E. and J.M.G. J.C.E. and J.M.G. performed the analyses. A.K. wrote the initial draft of the manuscript, J.M.G. made major editorial adjustments and A.K., C.J.E. and J.M.G. made further editorial adjustments; all authors approved the final version.
References
1. De Villiers CC, Pama C. Geslagsregisters van die ou Kaapse families,1 A-K [Genealogies of old Cape families]. Cape Town: AA Balkema; 1966. Afrikaans. [ Links ]
2. Ridley M. Evolution. Oxford: Blackwell Science Ltd; 2004. [ Links ]
3. Botha MC, Beighton PB. Inherited disorders in the Afrikaner population of southern Africa. Part I. Historical and demographic background, cardiovascular, neurological, metabolic and intestinal conditions. S Afr Med J. 1983;64:609-612. [ Links ]
4. Botha MC, Beighton PB. Inherited disorders in the Afrikaner population of southern Africa. Part II. Skeletal, dermal and haematological conditions; the Afrikaners of Gamkaskloof; demographic considerations. S Afr Med J.1983;64:664-667. [ Links ]
5. Nurse GT, Weiner JS, Jenkins T. The peoples of southern Africa and their affinities. Oxford: Oxford University Press; 1985. [ Links ]
6. Heese JA. Die herkoms van die Afrikaner 1657-1867 [Ancestry of the Afrikaner 1657-1867]. Cape Town: AA Balkema; 1971. Afrikaans. [ Links ]
7. Heese HF. Groep sonder grense [Group without borders]. Pretoria: Protea Boekhuis; 2005. Afrikaans. [ Links ]
8. De Bruyn GFC. Die samestelling van die Afrikanervolk [The composition of the Afrikaner population]. Tydskr Geesteswet. 1976;15:39-42. Afrikaans. [ Links ]
9. Greeff JM. Deconstructing Jaco: Genetic heritage of an Afrikaner. Ann Hum Genet. 2007;71:674-688. http://dx.doi.org/10.1111/j.1469-1809.2007.00363.x [ Links ]
10. Botha MC, Pritchard J. Blood group gene frequencies - An indication of the genetic constitution of population samples in Cape Town. S Afr Med J. 1972;46:S1-S27. [ Links ]
11. Eisenberger NI, Way BM, Taylor SE, Welch WT, Lieberman MD. Understanding genetic risk for aggression: Clues from the brain's response to social exclusion. Biol Psychiatry. 2007;61:1100-1108. http://dx.doi.org/10.1016/j.biopsych.2006.08.007 [ Links ]
12. Gallardo-Pujol D, Andres-Pueyo A, Maydeu-Olivares A. MAOA genotype, social exclusion and aggression: An experimental test of a gene-environment interaction. Genes Brain Behav. 2013;12:140-145. http://dx.doi.org/10.1111/j.1601-183X.2012.00868.x [ Links ]
13. Kuepper Y Grant P Wielpuetz C, Hennig J. MAOA-uVNTR genotype predicts interindividual differences in experimental aggressiveness as a function of the degree of provocation. Behav Brain Res. 2013;247:73-78. http://dx.doi.org/10.1016/j.bbr.2013.03.002 [ Links ]
14. Lea R, Chambers G. Monoamine oxidase, addiction, and the "warrior" gene hypothesis. N Z Med J. 2007;120:u2441. [ Links ]
15. Lan NC, Heinzmann C, Gal A, Klisak I, Orth U, Lai E, et al. Human monoamine oxidase A and B genes map to Xp 11.23 and are deleted in a patient with Norrie disease. Genomics. 1989;4:552-559. http://dx.doi.org/10.1016/0888-7543(89)90279-6 [ Links ]
16. Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273-279. http://dx.doi.org/10.1007/s004390050816 [ Links ]
17. Reti IM, Xu JZ, Yanofski J, Mckibben J, Uhart M, Cheng Y-J, et al. Monoamine oxidase A regulates antisocial personality in whites with no history of physical abuse. Compr Psychiatry. 2011;52:188-194. http://dx.doi.org/10.1016/j.comppsych.2010.05.005 [ Links ]
18. Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851-854. http://dx.doi.org/10.1126/science.1072290 [ Links ]
19. Greeff JM, Greeff FA, Greeff AS, Rinken L, Welgemoed DJ, Harris Y Low nonpaternity rate in an old Afrikaner family. Evol Hum Behav. 2012;33:268-273. http://dx.doi.org/10.1016/j.evolhumbehav.2011.10.004 [ Links ]
20. Greeff JM, Erasmus JC. Three hundred years of low non-paternity in a human population. Heredity. Forthcoming 2015. http://dx.doi.org/10.1038/hdy.2015.36 [ Links ]
21. Greeff JM, Erasmus JC. Appel Botha Cornelitz: The abc of a three hundred year old divorce case. Forensic Sci Int Genet. 2013;7:550-554. http://dx.doi.org/10.1016/j.fsigen.2013.06.008 [ Links ]
22. Giliomee H. Die Afrikaners: 'n Biografie [The Afrikaners: Biography of a people]. Cape Town: Tafelberg Uitgewers; 2004. Afrikaans. [ Links ]
23. Strassmann BI, Kurapati NT, Hug BF, Burke EE, Gillespie BW, Karafet TM, et al. Religion as a means to assure paternity. Proc Natl Acad Sci USA. 2012;109:9781-9785. http://dx.doi.org/10.1073/pnas.1110442109 [ Links ]
24. Prichard ZM, Mackinnon AJ, Jorm AF, Easteal S. AVPR1A and OXTR polymorphisms are associated with sexual and reproductive behavioral phenotypes in humans. Hum Mutat. 2007;28:1150. http://dx.doi.org/10.1002/humu.9510 [ Links ]
25. Walum H, Westberg L, Henningsson S, Neiderhiser JM, Reiss D, Igl W, et al. Genetic variation in the vasopressin receptor 1a gene (AVPR1A) associates with pair-bonding behavior in humans. Proc Natl Acad Sci USA. 2008;105:14153-14156. http://dx.doi.org/10.1073/pnas.0803081105 [ Links ]
26. Thibonnier M, Graves MK, Wagner MS, Chatelain N, Soubrier F, Corvol P et al. Study of V1-vascular vasopressin receptor gene microsatellite polymorphisms in human essential hypertension. J Mol Cell Cardiol. 200;32:557-564. http://dx.doi.org/10.1006/jmcc.2000.1108 [ Links ]
27. Hammock EAD, Young LJ. Variation in the vasopressin V1a receptor promoter and expression: Implications for inter- and intraspecific variation in social behaviour. Eur J Neurosci. 2002;16:399-402. http://dx.doi.org/10.1046/j.1460-9568.2002.02083.x [ Links ]
28. Meyer-Lindenberg A, Kolachana B, Gold B, Olsh A, Nicodermus KK, Mattay V et al. Genetic variants in AVPR1A linked to autism predict amygdala activation and personality traits in healthy humans. Mol Psychiatry. 2009;14:968-975. http://dx.doi.org/10.1038/mp.2008.54 [ Links ]
29. Knafo A, Israel S, Darvasi A, Bachner-Melman R, Uzefovsky F, Cohen L, et al. Individual differences in allocation of funds in the dictator game associated with length of the arginine vasopressin 1a receptor RS3 promoter region and correlation between RS3 length and hippocampal mRNA. Genes Brain Behav. 2008;7:266-275. http://dx.doi.org/10.1111/j.1601-183X.2007.00341.x [ Links ]
30. Avinum R, Israel S, Shalev I, Gritsenko I, Bornstein G, Ebstein RP et al. AVPR1A variant associated with preschoolers' lower altruism behaviour. PLoS One. 2011;6:e25274. http://dx.doi.org/10.1371/journal.pone.0025274 [ Links ]
31. Bachner-Melman R, Zohar AH, Bacon-Shnoor N, Elizur Y Nemanov L, Gritsenko I, et al. Link between vasopressin receptor AVPR1A promoter region microsatellites and measures of social behaviour in humans. J Indiv Diff. 2005;26:2-10. http://dx.doi.org/10.1027/1614-0001.26.L2 [ Links ]
32. Kim SJ, Young LJ, Gonen D, Veenstra-vanderWeele J, Courchesne R, Lord C, et al. Transmission disequilibrium testing of arginine vasopressin receptor 1a AVPR1A) polymorphisms in autism. Mol Psychiatry. 2002;7:503-507. http://dx.doi.org/10.1038/sj.mp.4001125 [ Links ]
33. Yirmiya N, Rosenberg C, Levi S, Salomon S, Shulman C, Nemanov L, et al. Association between the arginine vasopressin 1a receptor (AVPR1a) gene and autism in a family-based study: Mediation by socialization skills. Mol Psychiatry. 2006;11:488-494. http://dx.doi.org/10.1038/sj.mp.4001812 [ Links ]
34. Yang SY, Cho S-C, Yoo HJ, Cho IH, Park M, Yoe J, et al. Family-based association study of microsatellites in the 5' flanking region of AVPR1A with autism spectrum disorder in the Korean population. Psychiatry Res. 2010;178:199-201. http://dx.doi.org/10.1016/j.psychres.2009.11.007 [ Links ]
35. Granot RY, Frankel Y Gritsenko V Lerer E, Gritsenko I, Bachner-Melman R, et al. Provisional evidence that the arginine vasopressin 1a receptor gene is associated with musical memory. Evol Hum Behav. 2007;28:313-318. http://dx.doi.org/10.1016/j.evolhumbehav.2007.05.003 [ Links ]
36. Ukkola-Vuoti L, Oikkonen J, Onkamo P Karma K, Raijas P Jârvelâ I. Association of the arginine vasopressin receptor 1A (AVPR1A) haplotypes with listening to music. J Hum Genet. 2011;56:324-329. http://dx.doi.org/10.1038/jhg.2011.13 [ Links ]
37. Morley AP Narayanan M, Mines R, Molokhia A, Baxter S, Craig G, et al. AVPR1a and SLC6A4 polymorphisms in choral singers and non-musicians: A gene association study. PloS One. 2012;7:e31763. http://dx.doi.org/10.1371/journal.pone.0031763 [ Links ]
38. Bachner-Melman R, Dina C, Zohar AH, Constantini N, Lerer E, Hoch S, et al. AVPR1a and SLC6A4 gene polymorphisms are associated with creative dance performance. PloS Genet. 2005;1:e42. http://dx.doi.org/10.1371/journal.pgen.0010042 [ Links ]
39. Bachner-Melman R, Zohar AH, Elizur Y Nemanov L, Gritsenko I, Konis D, et al. Association between a vasopressin receptor AVPR1a promoter region microsatellite and eating behaviour measured by a self-report questionnaire (Eating Attitudes Test) in a family-based study of a nonclinical population. Int J Eat Dis. 2004;36:451-460. http://dx.doi.org/10.1002/eat.20049 [ Links ]
40. Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-98. [ Links ]
41. Pritchard Z, Easteal S. Characterization of simple sequence repeat variants linked to candidate genes for behavioural phenotypes. Hum Mut. 2006;27:120-126. http://dx.doi.org/10.1002/humu.9394 [ Links ]
42. Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47-50. [ Links ]
43. R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [ Links ]
44. Deckert J, Catalano M, Syagailo YV Bosi M, Okladnova O, Di Bella D, et al. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet. 1999;8:621-624. http://dx.doi.org/10.1093/hmg/8.4.621 [ Links ]
45. Lu R-B, Lee J-F, Ko H-C, Lin W-W, Chen K, Shih JC. No association of the MAOA gene with alcoholism among Han Chinese males in Taiwan. Prog Neuro-Psychopharmacol Biol Psychiatry. 2002;26:457-461. http://dx.doi.org/10.1016/S0278-5846(01)00288-3 [ Links ]
 Correspondence:
Correspondence:
Jaco Greeff
Department of Genetics
University of Pretoria, Private Bag
X20, Hatfield 0028
South Africa
Email: jaco.greeff@up.ac.za
Received: 20 Feb. 2015
Revised: 23 Apr. 2015
Accepted: 01 June 2015
Note: This article is supplemented with online only material.




![Dichloro(bis[diphenylthiourea])cadmium complex as a precursor for HDA-capped CdS nanoparticles and their solubility in water](/img/en/prev.gif)