Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.110 n.11-12 Pretoria Nov./Dec. 2014
http://dx.doi.org/10.1590/sajs.2014/20130306
RESEARCH ARTICLE
http://dx.doi.org/10.1590/sajs.2014/20130306
Biofilm formation in surface and drinking water distribution systems in Mafikeng, South Africa
Suma George MulamattathilI,II; Carlos BezuidenhoutI; Moses MbeweII
ISchool of Environmental Science and Development, North-West University,Potchefstroom, South Africa
IIDepartment of Water and Sanitation, University of Limpopo, Polokwane, South Africa
ABSTRACT
Poor quality source water and poorly treated reused wastewater may result in poor quality drinking water that has a higher potential to form biofilms. A biofilm is a group of microorganisms which adhere to a surface. We investigated biofilm growth in the drinking water distribution systems in the Mafikeng area, in the NorthWest Province of South Africa. Analysis was conducted to determine the presence of faecal coliforms, total coliforms, Pseudomonas spp. and Aeromonas spp. in the biofilms. Biofilms were grown on a device that contained copper and galvanised steel coupons. A mini tap filter - a point-of-use treatment device which can be used at a single faucet - was also used to collect samples. Scanning electron microscopy demonstrated that multi-species biofilms developed on all the coupons as well as on the point-of-use filters. Galvanised steel and carbon filters had the highest density of biofilm. Total coliforms, faecal coliforms and Pseudomonas spp. were isolated from raw water biofilm coupons only. Aeromonas spp. and Pseudomonas spp. were isolated from filters. The susceptibility of selected isolates was tested against 11 antibiotics of clinical interest. The most prevalent antibiotic resistance phenotype observed was KF-AP-C-E-OT-K-TM-A. The presence of virulence genes was determined using the polymerase chain reaction. These results indicate that bacteria present in the water have the ability to colonise as biofilms and drinking water biofilms may be a reservoir for opportunistic bacteria including Pseudomonas and Aeromonas species.
Keywords: Aeromonas; biofilm; drinking water distribution system; Pseudomonas; total coliforms
Introduction
Water is a vital resource for life and access to safe drinking water is a basic right of every individual.1 South Africa is a semi-arid country with very little rainfall, resulting in high water stress; as such, individuals in many communities struggle to access potable water.2 Water scarcity problems can be addressed through the recycling of municipal wastewater for reuse in households - a practice which is increasing worldwide.3 However, reclaimed water may be a major source of pathogenic and opportunistic microorganisms, as well as pharmaceutical waste products.4 The presence of pathogenic microorganisms in treated water sources usually is because they are able to survive the treatment process. Moreover, in most developing countries, water-treatment plants are usually faced with maintenance problems and a lack of qualified personnel.
In aquatic environments, microorganisms have the ability to adhere to solid surfaces and form biofilms.5 Biofilms are bacterial communities embedded in a polysaccharide matrix, which gives them the opportunity to resist destruction by antibiotics, environmental stress, biocides and detergents.6 Bacterial regrowth in the distribution system may result from the detachment of biofilm bacteria, which increases the risk of infection in humans when the water is consumed.6 Generally, most water distribution systems are characterised by the presence of biofilms, regardless of purity, the type of pipe material used for distribution or the presence of a disinfectant.7 Bacteria in drinking water systems can therefore grow in bulk water and as biofilms attached to the walls of pipes.8 Moreover, the development of biofilms inside water distribution pipes facilitates the propagation of mixed microbial populations and is considered the main source of planktonic bacteria in water supply systems.9 This problem is further aggravated by the presence of opportunistic pathogens such as Pseudomonas, Aeromonas, Klebsiella, Mycobacter, Escherichia coli, Helicobacter, Salmonella and Legionella spp. that may increase the health risks associated with the consumption of water from these sources.10
Different materials - such as cast iron galvanised steel, stainless steel, copper and polyethylene - have been used to manufacture water distribution pipes and these materials favour biofilm formation in the water distribution systems.7,11,12 Differences in the pipe materials greatly favour the survival of different bacterial species.13 The presence of biofilms in drinking water distribution pipes usually leads to a number of undesirable effects on the quality of water that is supplied to consumers.14 The development of biofilms in copper pipes facilitates cuprosolvency which increases the release of copper into the distribution system.15 Furthermore, increased carbon influences the growth of heterotrophic plate count bacteria which are also involved in the corrosion of copper.15 The corrosion of lead-containing plumbing materials increases the chances of lead contamination in tap water, which can cause adverse health effects in humans, especially children.16 Detachment of bacteria from the biofilms may affect the quality of the water.17 Therefore, the deterioration of the quality of drinking water as a result of biofilm formation is a major concern for most municipal supply agencies and communities. Biofilms are present in spite of different treatment processes; the occurrence of biofilms in drinking water is attributed to bacterial resistance to disinfectants, ability of the bacterial species to resist chemical compounds released from pipe materials and species association which increases the proportion of viable cells.18,19 Moreover, the use of different disinfectant methods may have long-term effects on the biofilm community.20 Biofilms consisting of Pseudomonas aeruginosa and different faecal bacterial species have been detected in water distribution systems, even in countries that have more advanced water-treatment facilities.21,22
Mafikeng is the capital of the North West Province of South Africa. This city uses both groundwater as well as dam water for drinking water production. Some areas receive a mixture of the two water types and others receive only one or the other. The purification plant for the surface water is at the Modimola Dam and receives treated wastewater. The water purification plant is downstream from the sewage treatment plant and is thus a semi-closed water conservation system.
The study was designed to investigate the biofilm forming ability, antibiotic resistance and virulence gene determinants of biofilm bacteria, especially Pseudomonas and Aeromonas species, in the water distribution systems in Mafikeng.
Materials and methods
Sampling area
Biofilm forming devices were installed at different sites within the water distribution system in the Mafikeng-Mmabatho area: (1) raw untreated water from the Modimola Dam, (2) treated water from household taps, received from the Modimola Dam treatment plant, (3) treated water from Molopo Eye, a natural spring and (4) water from the Modimola Dam treatment plant mixed with chlorinated water from Molopo Eye (mixed water).
Biofilm formation devices
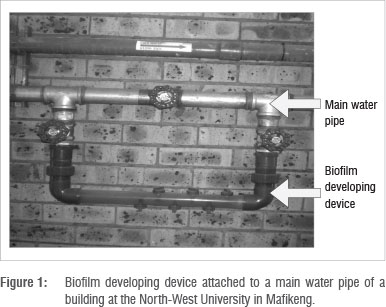
To study biofilm growth, a flow system technique that utilises a biofilm developing device was used (Figure 1). The biofilm developing device pipe system was made from clear plastic pipe with a diameter of 16 mm. The device was installed with copper and galvanised coupons to serve as solid surfaces onto which bacteria could adhere and form biofilms. Coupons were held in place by screws. The device was mounted horizontally to the main pipe of a building of the North-West University in Mafikeng (Figure 1). This building receives mixed water. The coupons were installed at the different sampling points for 6 months. Mini tap filters (Figure 2) - which are point-of-use (POU) treatment devices which can be used at a single faucet under constant flow - were also used to form biofilms during the second collection. The filters were placed on cold water taps in participating locations that received treated groundwater (Molopo Eye water), only Modimola Dam water or mixed water (North-West University, Mafikeng campus). The filters remained at these sampling points for 4 months.

Sampling of biofilm
Biofilm samples were analysed twice during the study period. To collect samples, the pipes containing biofilm developing devices were closed with valves before they were disconnected and the coupons were removed using sterile forceps. The filters in the mini tap filter devices were aseptically removed from the cartridge. Coupons were placed immediately into sterile 100-mL Schott bottles that contained water from the particular sampling site. Samples were transported on ice to the laboratory for analysis. Upon arrival in the laboratory, the coupons were removed from the bottles. Those coupons intended for scanning electron microscopy were stored in 100% alcohol and the remaining coupons were analysed for bacterial growth.
Scanning electron microscopy
The biofilm structure was investigated using scanning electron microscopy (SEM). Biofilm samples were fixed by 2.5% glutaraldehyde and 2% osmium tetroxide, dehydrated sequentially in increasing concentrations of ethanol (70%, 90% and 100%) for 15 min and critically dried in liquid carbon dioxide. The samples were mounted on SEM stubs using double-sided carbon tape. These stubs were then carbon coated. Finally, the samples were super coated with gold/palladium and viewed using a Philips XL30 scanning electron microscope (Philips, Aachen, Germany). Enlargements ranged from 63X to 20 000X.
Isolation of bacteria from the biofilm
Bacteria were isolated using standard methods. Biofilm bacteria on the coupons were removed by swabbing the surfaces with a sterile cotton swab dipped in sterile nutrient broth and immediately streaking it onto selective media for the isolation of targeted organisms. The media used were: mFC agar for the isolation of faecal coliforms, mEndo for total coliforms and Aeromonas selective agar for Aeromonas and Pseudomonas species. All the media used were Biolab agars from Merck (Johannesburg, South Africa). The plates were incubated aerobically at 35 °C for 24 h, except for mFC agar plates which were incubated at 45 °C for 24 h. Blue colonies from mFC agar and metallic-sheen colonies from mEndo agar were considered as presumptive faecal coliforms and total coliforms, respectively. Moreover, yellow and green colonies on Aeromonas selective agar represented Aeromonas and Pseudomonas species, respectively. These isolates were sub-cultured on the respective selective media and plates were incubated aerobically at 35 °C and 45 °C, respectively, for 24 h. Presumptive pure colonies were subjected to specific preliminary and confirmatory biochemical tests. All pure isolates were Gram stained using standard methods.
Preliminary biochemical tests
Triple sugar iron agar test
The triple sugar iron (TSI) test can also be used as a confirmation test for E. coli. The TSI agar obtained from Biolab (Merck, Johannesburg, South Africa) was used to determine the ability of isolated organisms to utilise the substrates glucose, sucrose and lactose at sample concentrations of 0.1%, 1.0% and 1.0%, respectively. The test was performed as per the instructions of the manufacturer and evaluated based on the formation of gas and hydrogen sulphide and fermentation of carbohydrates to produce acids. TSI agar slants were prepared in sterile 15-mL McCartney bottles. The slants were streak and stab inoculated with a sterile inoculation needle containing the selected colony. Following inoculation, the slants were incubated at 37 °C for 24 h. The colour of the slant and butt was recorded (red or yellow) as well as the production of hydrogen sulphide (when the agar blackened) and formation of gas (when the agar split).
Oxidase test
This test was performed using the Test Oxidase™ reagent (PL.390) from Mast Diagnostics (Nesto, Wirral, UK) in accordance with the manufacturer's published protocol. A well-isolated pure colony was placed on filter paper using a sterile wire loop. A drop of Test OxidaseTM reagent was added to the filter paper and mixed. After 30 s the filter was observed for a colour change. Isolates that produced a purple colour were presumptively considered to be E. coli. Oxidase positive colonies were taken as presumptive Aeromonas isolates.
Analytical profile index test
The analytical profile index (API) 20E test was performed in accordance with the manufacturer's protocol (BioMeriéux, Marcy I'Etoile, France). An API 20E strip was used to identify Enterobacteriaceae and other non-fastidious, Gram-negative rods. The strips were inoculated and incubated at 37 °C for 24 h. The indices obtained after reading the results were interpreted using the API web software (BioMeriéux®). The organisms were identified to species level.
Antimicrobial susceptibility test
An antibiotic susceptibility test was performed using the Kirby-Bauer disc diffusion method. The following antibiotic discs (Mast Diagnostics, UK) were used at the final concentrations that are indicated: ampicillin (AP) 10 µg, cephalothin (KF) 5 µg, streptomycin (S) 10 µg, erythromycin (E) 15 µg, chloramphenicol (C) 30 µg, neomycin (NE) 30 µg, amoxycillin (A) 10 µg, ciprofloxacin (CIP) 5 µg, trimethoprim (TM) 25 µg, kanamycin (K) 30 µg, oxytetracycline (OT) 30 µg. These antibiotics were chosen because either they are used in both humans and other animals or they have been reported to be resistant in previous studies.
Three colonies were picked from each sample and each colony was transferred into 3 mL of sterile distilled water to prepare a bacterial suspension. Aliquots of 1 mL from each suspension were spread plated on Mueller-Hinton agar plates. Antibiotic discs were applied to the plates using sterile needles and the plates were incubated at 37 °C for 24 h. After incubation, the antibiotic inhibition zone diameters were measured. Results obtained were used to classify isolates as being resistant, intermediate resistant or susceptible to a particular antibiotic using standard reference values according to the US National Committee for Clinical Laboratory Standards (now called the Clinical and Laboratory Standards Institute). Multiple antibiotic resistance phenotypes were generated for isolates that showed resistance to three or more antibiotics.
Confirmatory DNA test
Genomic DNA extraction
Genomic DNA was extracted from all the presumptive Pseudomonas and Aeromonas isolates using the alkaline lysis method. The concentration and quality of the extracted DNA in solution were determined using a spectrophotometer (NanoDrop ND 1000, Thermo Scientific, USA) and 1% (w/v) agarose gel electrophoresis. The latter was also used for determining the integrity of the genomic DNA.
Polymerase chain reaction for identifying species
The identities of the presumptive Pseudomonas species were confirmed through amplification of the toxA24 and ecfX25 gene determinants and Aeromonas species were confirmed through gyrB23 amplification. Polymerase chain reactions (PCRs) were performed using oligonucleotide primer combinations under the cycling conditions given in Table 1. Standard 25-µL reactions that consisted of 1 µg/uL of the template DNA, 50 pmol of each oligonucleotide primer set, 1X PCR master mix and RNase free water were prepared. Amplifications were performed using a Peltier Thermal Cycler (model-PTC-220DYAD™ DNA ENGINE, MJ Research Inc., Waltham, MA, USA). All PCR reagents used were Fermentas (USA) products supplied by Inqaba Biotec Pty Ltd (Pretoria, South Africa). PCR products were subjected to 1% (w/v) agarose gel electrophoresis.
Polymerase chain reaction for detection of virulence gene markers
Pseudomonas species were screened for the presence of the exoA, exoS and exoT virulence gene determinants26 while a specific PCR for the detection of aerA and hylH genes was performed on all positively identified Aeromonas species27. PCRs were performed using oligonucleotide primer combinations under the cycling conditions given in Table 2. Amplifications were performed using a Peltier Thermal Cycler (model-PTC-220DYAD™ DNA ENGINE). The reactions were prepared in 25-µL volumes that constituted 1 µg/µL of the template DNA, 50 pmol of each oligonucleotide primer set, 1X PCR master mix and RNase free water. All PCR reagents used were Fermentas (USA) products supplied by Inqaba Biotech Pty Ltd (Pretoria, South Africa). pCr products were subjected to 2% (w/v) agarose gel electrophoresis.
Electrophoresis of polymerase chain reaction products
Products of the PCRs were separated by electrophoresis on 2% (w/v) agarose gel. Electrophoresis was conducted in a horizontal Pharmacia Biotech equipment system (model Hoefer HE 99X; Amersham Pharmacia Biotech, Uppsala, Sweden) for 2 h at 60 V using 1X TAE buffer (40 mM Tris, 1 mM EDTA and 20 mM glacial acetic acid, pH 8.0). Each gel contained a 100-bp DNA molecular weight marker (Fermentas, Hanover,MD, USA). The gels were stained in ethidium bromide (0.1 ^g/ml) for 15 min and amplicons were visualised under UV light. A Gene Genius Bio imaging system (Syngene, Synoptics, Cambridge, UK) was used to capture the image using GeneSnap (version 3.07.01) software (Syngene, Synoptics) to determine the relative size of amplicons.
Results
Occurrence and diversity of microorganisms in the bio films
Table 3 indicates the different types of organisms isolated from the biofilm. The results indicate only the numbers of isolates that were positive for the various categories when subjected to preliminary tests (TSI, oxidase and, in the case of Pseudomonas spp. and Aeromonas spp., API 20E). It is evident from Table 3 that total coliforms and faecal coliforms were present in the biofilms from the raw water from Modimola Dam but were not detected in the biofilms of the treated water from Modimola Dam. Furthermore, Aeromonas and Pseudomonas spp. were detected in the biofilms of the raw dam water as well as the drinking water from the POU devices. Only Pseudomonas spp. (27) were isolated from the biofilm of the Modimola Dam raw water (Table 3), whereas both Aeromonas spp. and Pseudomonas spp. were found in the biofilms of the treated Modimola Dam drinking water and the mixed water.
Figures 3 to 5 depict the surfaces of coupons and filters exposed to raw water (Figure 3) and treated water distribution systems (Figures 4 and 5). Different organisms were isolated from the metal surfaces. The SEM revealed the existence of bacterial cells within the biofilm matrix, with a greater amount of bacterial cells found on the galvanised coupons than on the copper coupons (Figures 3 to 5). From these micrographs, it was also evident that high bacterial densities were found in biofilms from the raw water of the Modimola Dam as well as from the drinking water distribution system in Mafikeng. The dam water is treated and supplied to homes for consumption.
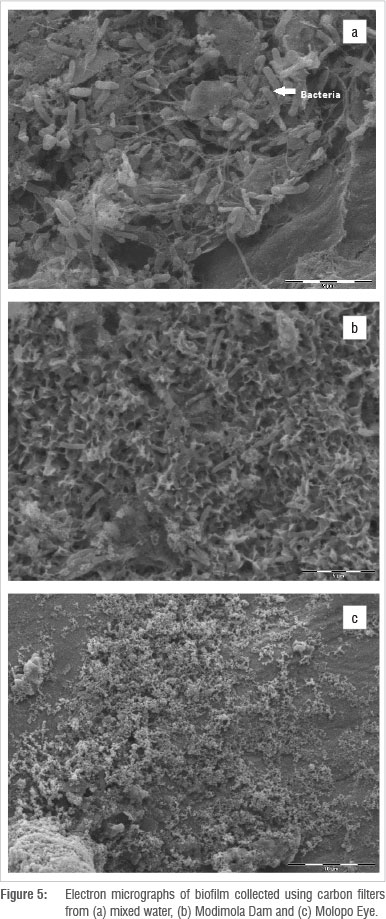
When the coupons were removed from the biofilm development device, a green colour was noticed on the copper coupons; this colour is attributed to the corrosion products that are visible (like crystals) in the SEM (Figure 4b). Attached cells in association with exopolysaccharide were visible in galvanised, copper and POU filter surfaces in the SEMs (Figures 3 to 5). It is therefore suggested that the suspended cells have the potential to attach to the surface and participate during biofilm formation. An extensive sponge-like exopolysaccharide layer can be seen on the galvanised coupons in association with bacterial cells (Figure 3a). Figure 5 (POU filters) shows evidence of biofilm formation on the surface of filters from mixed water, dam water and water from the Molopo Eye. Rod-shaped bacteria are the dominating organisms in the biofilm and the aggregation of rod-shaped bacteria entangled in the exopolysaccharide, as seen in the micrographs, usually reflect a mature biofilm. It was found that galvanised coupons from raw and treated water contained thicker biofilms than did the copper coupons. Of the treated water from the three sites, coupons from the mixed water were colonised by a variety of bacteria.
If biofilms contain any pathogenic bacteria, the detachment of biofilms could release these bacteria into drinking water and affect risk levels of consumers.7 In another study, coliform bacteria - originating from biofilms observed on rubber-coated valves - were isolated from drinking water distribution systems.21 Coliforms were not detected in the present study, but that does not mean that they were absent.
Antimicrobial susceptibility test
All the organisms were subjected to an antibiotic sensitivity test using 11 antibiotics of clinical importance. Multiple antibiotic resistance phenotypes were generated for isolates resistant to three or more drugs; the results are shown in Table 4.
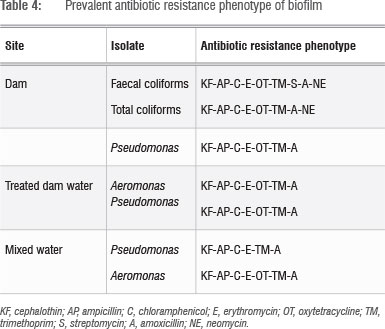
All the isolates tested were resistant to ampicillin, amoxicillin, cephalothin, erythromycin, chloramphenicol and trimethoprim. All the organisms tested were susceptible to ciprofloxacin. Four different multiple antibiotic resistance patterns were observed and all the isolates were resistant to three or more classes of antibiotics. The highest level of resistance -with the phenotype KF-AP-C-E-OT-K-TM-A, indicating resistance to eight drugs - was observed for isolates from biofilms. From these results, it is evident that ciprofloxacin and streptomycin were the most effective, because all or a large proportion of the isolates were susceptible to both. These results indicate that biofilm grown organisms may serve as a reservoir for antibiotic-resistant organisms, and therefore may have the potential to cause infections. This finding is a cause for concern, particularly for infants, the elderly and immunocompromised individuals in the Mafikeng community.
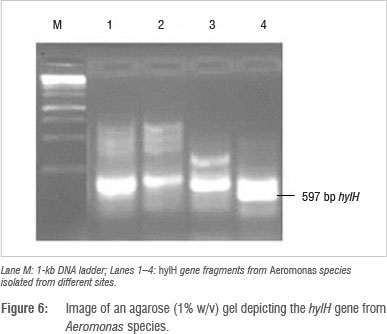
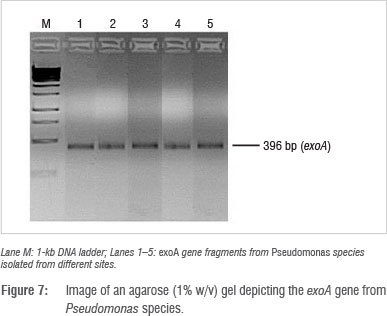
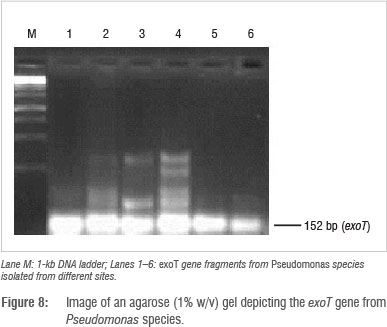
Identification and detection of virulence gene isolates were done using gyrB, toxA and ecfX gene fragments through PCR. The gyrB and ecfX gene fragments were amplified. Specific PCR assays for the detection of virulence genes (aerA and hylH in Aeromonas and exoA, exoS and exoT in Pseudomonas) produced DNA fragments of the expected size of some of the markers. Of the 39 isolates that were screened, the combination of virulence genes detected in the isolates from the different areas are shown in Table 5. Aeromonas spp. that were isolated from biofilms from Modimola Dam raw water (8 of 12) and mixed water (1 of 10) harboured the hylH gene (Figure 6). These genes were more prevalent in isolates from raw dam water. However, aerA genes were not detected in the isolates from either site. The exoA gene (Figure 7) was detected in Pseudomonas spp. from the raw water biofilm and biofilm isolates from the treated dam water. Isolates from the biofilms from all sites harboured exoT genes (Figure 8). However, none of the Pseudomonas spp. isolates possessed the exoS gene.
Discussion
Water is a vital component of life but can serve as an important vehicle for the dissemination of potential pathogens to humans.21 Source water that receives sewage effluent may be polluted with opportunistic pathogenic microorganisms and pharmaceuticals.28 Modimola Dam receives treated sewage effluent. Organisms that survived the treatment process may be able to grow in the aquatic environment. A further concern is that, at times, the treatment processes fail and the potential exists for the presence of opportunistic pathogens such as Aeromonas hydrophila and Pseudomonas aeruginosa. Both of these species have a tendency to form biofilms. This study was thus aimed at determining whether Aeromonas spp. and Pseudomonas spp. occur in biofilms in the drinking water of Mafikeng.
Organisms isolated in this study include faecal coliforms, total coliforms, Aeromonas and Pseudomonas. In a similar study, heterotrophic bacteria were isolated from biofilms in copper plumbing, which included a wide variety of organisms.10Aeromonas species are implicated in gastroenteritis and are generally considered to be waterborne pathogens29 while Pseudomonas species are opportunistic pathogens that cause nosocomial infections in susceptible patients30. Moreover, it is very difficult to eradicate Pseudomonas species because of their high intrinsic resistance to a variety of antibiotics, including ß-lactams, aminoglycosides and fluroquinolones.31Aeromonas has the potential to grow in water distribution systems, especially in biofilms, where it is resistant to chlorination and produces many different putative virulence factors.32 Isolates that are resistant to chlorine may be present in tap water that is intended for human consumption, and which therefore causes disease in humans.
The regrowth and formation of biofilms in drinking water distribution pipes has been detected even in countries with advanced water-treatment and health-care facilities.21 The presence of biofilms has been found to cause significant corrosion of pipe materials and, subsequently, the addition of inorganic and organic matter, which results in a poor aesthetic quality of water.12 Moreover, free chlorine reacts with compounds present in biofilms inside water pipes, producing an unpleasant taste and odour.11 Adherent bacteria are more resistant to antimicrobial agents and could contribute to the planktonic cells present in the bulk water. The prevalence of planktonic bacteria in drinking water may be a result of sloughing of the biofilm - an assumption which is supported by the observation of planktonic bacterial episodes in treated drinking water.5
Biofilm formation is facilitated by many factors including available nutrients, characteristics of the pipe material, disinfectants used, physico-chemical parameters and the ability of the microorganisms to resist destruction by antimicrobial agents.33 The significant release of nutrients from the surface material to the water promotes the growth of bacteria.18
Cementitious, metallic and plastic materials are the three most commonly used types of plumbing materials. The plumbing materials chosen for this study were copper and galvanised steel, which are commonly used in domestic plumbing systems in South Africa. Our results demonstrate biofilm formation on both of the plumbing materials that were used. In another study, a higher density of bacteria was observed on polyethylene and polyvinylchloride surfaces than on galvanised steel.34 In the present study, thicker biofilms were observed on galvanised steel than on copper. However, biofilms on galvanised steel coupons were not compared with those on polyethylene and polyvinylchloride surfaces. It has been observed that biofilms on copper had low concentrations of culturable bacteria.35 It is thus not uncommon to find low levels of culturable bacteria in biofilms on copper coupons. Moreover, it has been reported that the formation of biofilms was slower on copper pipes than on polyethylene pipes. However, after 200 days there was no difference in microbial numbers in biofilms between the two pipe materials.18 This finding implies that in the case of long-term use, biofilm levels on metal surfaces will be similar to those on polyethylene and polyvinylchloride surfaces. However, as a consequence of their nature, biofilms on metal surfaces will contribute to microbial-induced corrosion, which can increase the metal concentration in water distributed by copper pipes,15 which has the potential to cause health problems.
In the present study, we did not focus on the corrosion potential of the biofilms but rather on whether or not Pseudomonas and Aeromonas spp. colonise the biofilms. It has been previously demonstrated that Pseudomonas spp. are opportunistic pathogens that can integrate into drinking water biofilms on materials which are relevant for domestic plumbing systems.35 Biofilms have been implicated in human infections and are particularly recalcitrant to antibiotic compounds.30,36 Final water produced by the water-treatment plant must comply with the South African National Standard (SANS) 241:2011 for drinking water.37
No total coliforms or faecal coliforms were detected in biofilms from the treated drinking water, which thus complied with the SANS 241 standard of 0 CFU/100 mL. This finding is an indication that the treatment process was effective in removing total coliforms and faecal coliforms from the raw water. However, Pseudomonas and Aeromonas spp. were isolated from the biofilms of treated and filtered water. This may be an indication of deteriorating water quality. High levels of Pseudomonas spp. in water may cause taste, odour and turbidity problems.38 There is no available SANS 241 standard for Pseudomonas and Aeromonas spp. in drinking water.
Identities of the organisms were confirmed through PCR using gyrB, toxA, ecfX, aerA and hylH gene fragments. The ecfX gene encodes an extra cytoplasmic function sigma factor, which may be involved in haem uptake or virulence39, whereas the gyrB gene encodes the DNA gyrase subunit B, a protein which plays a crucial role in the DNA replication process40 and the toxA gene encodes the exotoxin A precursor25. PCR assays targeting the ecfX and gyrB genes are highly suitable for the identification of P. aeruginosa.25,39 Application of the PCR technique to target gyrB, aerA and hylH genes is an excellent molecular chronometer for screening potentially virulent Aeromonas species in food and the environment.4041
One rational approach to determine whether Pseudomonas and Aeromonas spp. have the potential to be virulent is the assessment of virulence phenotypes and screening for specific virulence genes. Pseudomonas and Aeromonas spp. isolated in the present study carried some gene sequences encoding toxic proteins, indicating the potential of these organisms to cause diseases in humans. The ability of Pseudomonas spp. to express these virulence determinants also enhances their capabilities to produce biofilms.36Pseudomonas aeruginosa is able to synthesise a large number of virulence proteins that greatly influence pathogenesis.26Pseudomonas species produce extracellular compounds which promote adhesion and the ability of the isolates to attach to surfaces, thereby also increasing the virulence properties. The pathogenicity of Aeromonas is complex and multifactorial and has been linked to exotoxins such as cytolytic enterotoxin, haemolysin/aerolysin, (aerA, hylH, hylA, alt and ast), lipase and protease and various other cell-associated factors.2732 Screening for specific cytotoxin and haemolysin genes appeared to be the most effective way of detecting and characterising Aeromonas virulence factors.41
The desired gene fragments were successfully amplified, which indicates the presence of virulent Pseudomonas and Aeromonas spp. in biofilms from drinking water. From the molecular data it was demonstrated that the exoA, exoT and hylH genes were successfully used for the detection of virulent Pseudomonas and Aeromonas spp. in raw and drinking water biofilm samples. It is thus important to perform molecular confirmation of isolates to ensure accurate results.
Detection of these genes amongst the Pseudomonas and Aeromonas spp. isolated from the drinking water sources of Mafikeng is cause for concern and should be further investigated. PCR assays could provide a powerful supplement to the conventional methods for a more accurate risk assessment and monitoring of potentially virulent Pseudomonas and Aeromonas spp. in the environment.
Conclusion
Bacterial biofilms were detected in all water sources that were sampled and opportunistic pathogens such as Pseudomonas and Aeromonas species were isolated from biofilms in raw water from the Modimola Dam, drinking water and mixed water. These isolates were found to harbour virulence gene determinants indicating that they have the potential to cause diseases in humans. Therefore, it is important to constantly determine the occurrence of these species in water bodies and drinking water distribution systems, in particular, and to determine whether conditions prevail that may allow these opportunistic species to survive water purification processes. Such a strategy will be of particular importance in scenarios in which treated wastewater is reused for drinking water. A clear understanding of the different mechanisms by which biofilm bacteria harbour and distribute virulence factors, as well as protect themselves from the action of disinfectants and antibiotics, is vital to formulate control and management strategies.
Acknowledgements
We thank the National Research Foundation of South Africa (FA20040101000030 and FA2006040700029) for financial support of this study. We also thank the staff of the microbiology laboratory at Animal Health, NWU (Mafikeng) for their assistance. We acknowledge the assistance received from Mrs Rika Huyser and the employees of Mmabatho water works.
Authors' contributions
S.G.M. is the main author of the article and performed all the research work. C.B. supervised the work and guided the main author and M.M. co-supervised the work.
References
1. World Health Organization (WHO). Emerging issues in water and infectious disease. Geneva: WHO; 2003. [ Links ]
2. Adewumi J, Ilemobade A, Van Zyl J. Treated wastewater reuse in South Africa: Overview, potential and challenges. Resour Conserv Recycl. 2010;55:221-231. http://dx.doi.org/10.1016/j.resconrec.2010.09.012 [ Links ]
3. Revit D, Eriksson E, Donner E. The implications of household grey water treatment and reuse for municipal wastewater flows and micro-pollutant loads. Water Res. 2011;45:1549-1560. http://dx.doi.org/10.1016/j.watres.2010.11.027 [ Links ]
4. Yi L, Jiao W, Chen X, Weiping Chen W. An overview of reclaimed water reuse in China. J Environ Sci. 2011;23(10):1585-1593. http://dx.doi.org/10.1016/S1001-0742(10)60627-4 [ Links ]
5. Castonguay M, Van der Schaaf S, Koester W, Krooneman J, Van der Meer W Landini HP Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res Microbiol. 2006;157:471-478. http://dx.doi.org/10.1016/j.resmic.2005.10.003 [ Links ]
6. Simpson D. Biofilm processes in biologically active carbon water purification. Water Res. 2008;42(12):2839-2848. http://dx.doi.org/10.1016/j.watres.2008.02.025 [ Links ]
7. Lehtola M, Laxander M, Miettinen I, Hirvonen A, Vartiainen T, Martikainen T. The effects of changing water flow velocity on the formation of biofilms and water quality in pilot distribution systems consisting of copper or polyethylene. Water Res. 2006;40:2151-2160. http://dx.doi.org/10.1016/j.watres.2006.04.010 [ Links ]
8. Srinivasan S, Harrington G, Xagoraraki I, Goel R. Factors affecting bulk to total bacteria ratio in drinking water distribution systems. Water Res. 2008;42:3393-3404. http://dx.doi.org/10.1016/j.watres.2008.04.025 [ Links ]
9. Momba M, Kfir R, Venter S, Cloete T. An overview of biofilm formation in distribution systems and its impact on the deterioration of water quality. Water SA. 2000;26:59-66. [ Links ]
10. Critchley MM, Cromar NJ, MtCiure NC, Fallowfield HJ. The influence of the chemical composition of drinking water on cuprosolvency by biofilm bacteria. J Appl Microbiol. 2003;94:501-507. http://dx.doi.org/10.1046/j.1365-2672.2003.01857.x [ Links ]
11. Skjevrak I, Lund V Ormerod K, Herikstad H. Volatile organic compounds in natural biofilm in polyethylene pipes supplied with lake water and treated water from the distribution network. Water Res. 2005;39:4133-4141. http://dx.doi.org/10.1016/j.watres.2005.07.033 [ Links ]
12. Teng F, Guan Y Zhu W. Effect of biofilm on cast iron pipe in drinking water distribution system: Corrosion scales characterization and microbial community structure investigation. Corrosion Sci. 2008;50:2816-2823. http://dx.doi.org/10.1016/j.corsci.2008.07.008 [ Links ]
13. Kalmbach S, Manz W, Bendinger B, Szewzyk U. In situ probing reveals Aquabacterium commune as a widespread and highly abundant bacterial species in drinking water biofilms. Water Res. 2000;34:575-581. http://dx.doi.org/10.1016/S0043-1354(99)00179-7 [ Links ]
14. Tien C, Wu W, Tzu-Liang Chuang T, Chen C. Development of river biofilms on artificial substrates and their potential for biomonitoring water quality. Chemosphere. 2009;76:1288-1295. http://dx.doi.org/10.1016/j.chemosphere.2009.06.013 [ Links ]
15. Roslev P Larsen M, Jorgensen D, Hesselsoe M. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J Microbiol Meth. 2004;59:381-393. http://dx.doi.org/10.1016/j.mimet.2004.08.002 [ Links ]
16. Goudier M, Bouzid J, Sayadi S, Montiel A. Impact of orthophosphate addition on biofilm development in drinking water distribution systems. J Hazard Mater. 2009;167:1198-1202. http://dx.doi.org/10.1016/j.jhazmat.2009.01.128 [ Links ]
17. Elenter D, Milferstedt K, Zhang W, Hausner M, Morgenroth E. Influence of detachment on substrate removal and microbial ecology in a heterotrophic/ autotrophic biofilm. Water Res. 2007;41:4657-4671. http://dx.doi.org/10.1016/j.watres.2007.06.050 [ Links ]
18. Lehtola MJ, Miettinena IT, Keinanen MM, Kekkia TK, Laineb O, Hirvonen A. Microbiology, chemistry and biofilm development in a pilot drinking water distribution system with copper and plastic pipes. Water Res. 2004;38:p.3769-3779. http://dx.doi.org/10.1016/j.watres.2004.06.024 [ Links ]
19. Simöes M, Simöes L, Vieira M. Species association increases biofilm resistance to chemical and mechanical treatments. Water Res. 2009;43:229-237. http://dx.doi.org/10.1016/j.watres.2008.10.010 [ Links ]
20. Roeder R, Lenz J, Tarne P, Gebel J, Exner M, Szewzyk U. Long-term effects of disinfectants on the community composition of drinking water biofilms. Int J Hyg Environ Health. 2010;213:183-189. http://dx.doi.org/10.1016/j.ijheh.2010.04.007 [ Links ]
21. Kilb B, Lange B, Schaule G, Flemming H, Wingender J. Contamination of drinking water by coliforms from biofilms grown on rubber-coated valves. Int J Hyg Environ Health. 2003;206:563-573. [ Links ]
22. Werner E, Roe F, Bugnicourt A, Franklin M, Haydorn A, Molin S. Stratified gowth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2004;70(10):6188-6196. http://dx.doi.org/10.1128/AEM.70.10.6188-6196.2004 [ Links ]
23. Qin X, Emerson J, Stapp J, Stapp L, Abe P Burns J. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting Gram-negative bacilli from patients with cystic fibrosis. J Clin Microbiol. 2003;41:4312-4317. http://dx.doi.org/10.1128/JCM.41.9.4312-4317.2003 [ Links ]
24. Khan A, Cerniglia C. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl Environ Microbiol. 1994;60:3739-3745. [ Links ]
25. Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species specific ecfX gene target. J Microbiol Methods. 2007;70:20-29. http://dx.doi.org/10.1016/j.mimet.2007.03.008 [ Links ]
26. Kaszab E, Szoboszlay S, Dobolyi C, Háhn J, Kriszt B. Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from compost. Bioresour Technol. 2011;102:1543-1548. http://dx.doi.org/10.1016/j.biortech.2010.08.027 [ Links ]
27. Yogananth N, Bhakyaraj R, Chanthuru A, Anbalagan T, MullaiNila K. Detection of virulence gene in Aeromons hydrophila isolated from fish samples using PCR technique. Global J Biotech Biochem. 2009;4:51-53. [ Links ]
28. Köck-Schulmeyer M, Ginebreda A, Postigo C, López-Serna R, Pérez S, Brix R. Wastewater reuse in Mediterranean semi-arid areas: The impact of discharges of tertiary treated sewage on the load of polar micro pollutants in the Llobregat River (NE Spain). Chemosphere. 2011;82:670-678. http://dx.doi.org/10.1016/j.chemosphere.2010.11.005 [ Links ]
29. Pablos M, Huys G, Cnockaert M, Rodriguez-Calleja J, Otero A, Santos J, et al. Identification and epidemiology relationships of Aeromonas isolates from patients with diarrhoea, drinking water and foods. Int J Food Microbiol. 2011;147:203-210. http://dx.doi.org/10.1016/jjjfoodmicro.2011.04.006 [ Links ]
30. Fricks-Lima J, Hendrickson C, Allegaier M, Zhuo H, Wiener-Kronish J, Lynch S, et al. Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. Int J Antimicrob Agents. 2011;37:309-315. http://dx.doi.org/10.1016/jjjantimicag.2010.12.017 [ Links ]
31. Bredenstein E, De la Fuente-Núnez C, Hancock R. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011;19:419-426. http://dx.doi.org/10.1016/j.tim.2011.04.005 [ Links ]
32. Pablos M, Rodriguez-Celleja J, Santos J, Garcia-Lopez M. Occurrence of motile Aeromonas in municipal drinking water and distribution of genes encoding virulence factors. Int J Food Microbiol. 2009;135:158-164. http://dx.doi.org/10.1016/j.ijfoodmicro.2009.08.020 [ Links ]
33. Manuel CM, Nunes O, Melo L. Dynamics of drinking water biofilm in flow conditions. Water Res. 2007;47:551-562. http://dx.doi.org/10.1016/j.watres.2006.11.007 [ Links ]
34. Cloete TE. Resistance mechanism of bacteria to antimicrobial compounds. Int Biodeterior Biodegrad. 2003;51:277-282. http://dx.doi.org/10.1016/S0964-8305(03)00042-8 [ Links ]
35. Moritz M, Flemming H, Wingender J. Integration of Pseudomonas aeruginosa and Legionella pneumophila in drinking water biofilms grown on domestic plumbing materials. Int J Hyg Environ Health. 2013;213:190-197. http://dx.doi.org/10.1016/j.ijheh.2010.05.003 [ Links ]
36. Pimenta A, Martino P, Bouder E, Hulen C, Blight M. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microb Infect. 2003;5:1177-1187. http://dx.doi.org/10.1016/j.micinf.2003.09.002 [ Links ]
37. Department of Water Affairs (DWA). Blue drop handbook version 1 [document on the Internet]. [ Links ] c2012 [cited 2014 Mar 23]. Available from: http://www.dwa.gov.za/dir_ws/DWQR/Subscr/ViewNewsDoc.asp?FileID=262
38. World Health Organization (WHO). Guidelines for drinking water. 4th ed. Geneva: WHO; 2011. p. 541. [ Links ]
39. Anuj S, Whiley D, Kidd T, Bell S, Wainwright C, Nissen M, et al. Identification of Pseudomonas aeruginosa by a duplex polymerase chain reaction assay targeting the ecfX and gyrB genes. Diagn Microbiol Infect Dis. 2009;63:127-131. http://dx.doi.org/10.1016/j.diagmicrobio.2008.09.018 [ Links ]
40. Yánez M, Valor C, Catalán V. A simple and cost-effective method for the quantification of total coliforms and Escherichia coli in potable water. J Microbiol Methods. 2006;65:608-611. http://dx.doi.org/10.1016/j.mimet.2005.09.005 [ Links ]
41. Yousr A, Napis S, Rusul G, Son R. Detection of aerolysin and hemolysin genes in Aeromonas spp. isolated from environmental and shellfish sources by polymerase chain reaction. ASEAN Food J. 2007;14(2):115-122. [ Links ]
 Correspondence:
Correspondence:
Suma George Mulamattathil
Department of Water and Sanitation
University of Limpopo
Polokwane campus
Private Bag X1106
Sovenga 0727
South Africa
EMAIL: sgmulamattathil@gmail.com
Received: 27 Sep. 2013
Revised: 09 Jan. 2014
Accepted: 13 Apr. 2014














