Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.110 no.9-10 Pretoria oct. 2014
http://dx.doi.org/10.1590/sajs.2014/20130347
RESEARCH ARTICLE
Escherichia coli with virulence factors and multidrug resistance in the Plankenburg River
Corne Lamprecht; Marco Romanis; Nicola Huisamen; Anneri Carinus; Nika Schoeman; Gunnar O. Sigge; Trevor J. Britz
Department of Food Science, Stellenbosch University, Stellenbosch, South Africa
ABSTRACT
Escherichia coli is a natural inhabitant of the gut and E. coli levels in water are considered internationally to be an indication of faecal contamination. Although not usually pathogenic, E. coli has been linked to numerous foodborne disease outbreaks, especially those associated with fresh produce. One of the most common ways through which E. coli can be transferred onto fresh produce is if contaminated water is used for irrigation. In this study, a total of 81 confirmed E. coli strains were isolated from the Plankenburg River as part of three separate studies over 3 years. During sampling, E. coli levels in the river were above the accepted levels set by the World Health Organization and the South African Department of Water Affairs and Forestry for safe irrigation of fresh produce, which indicates that transfer of E. coli during irrigation is highly probable. Multiplex polymerase chain reaction screening for pathogenic gene sequences revealed one enteroaggregative positive strain and four enteropathogenic positive strains. The four enteropathogenic strains were also found to be resistant to three or more critically and highly important antibiotics and were therefore classified as multidrug resistant strains. These results show that E. coli with enteropathogenic potential and multiple antimicrobial resistance properties has persisted over time in the Plankenburg River.
Keywords: enteropathogenic; enteroaggregative; virulence genes; antibiotic resistance; pollution.
Introduction
Escherichia coli is a natural inhabitant of the gut of humans, birds and other warm-blooded animals and is widely accepted as an indicator of faecal contamination of water. It is a robust bacterium which is genetically highly adaptable to environmental stresses, and has been shown to survive and multiply in the environment.1,2 Based on the aforementioned, concerns have been raised in recent years regarding the status of E. coli as just a faecal indicator organism.3
Although most strains are commensal, pathogenic E. coli strains can contain various virulence factors and can be responsible for a variety of infections.4 Based on the specific virulence factors present, pathogenic E. coli can be classified as either extra-intestinal pathogenic E. coli (ExPEC) or intestinal pathogenic E. coli (InPEC). ExPEC strains are usually able to cause infections in anatomical sites outside of the intestinal tract and are associated with urinary tract infections, neonatal meningitis and septicaemia. ExPEC, like commensal E. coli, can colonise the intestinal tract without causing gastroenteritis. In contrast, InPEC strains can cause different types of gastroenteritis and can be divided into six pathogenic groups: enterohaemorrhagic (EHEC); enteropathogenic (EPEC); enteroaggregative (EAEC); enterotoxigenic (ETEC); enteroinvasive (EIEC) and diffusely adherent (DAEC) E. coli. Each of the InPEC types has different infection mechanisms and symptoms.4
Foodborne disease outbreaks linked to pathogenic E. coli, specifically those derived from fresh produce, are increasing both in number and intensity.5 As a result, E. coli is considered to be an emerging pathogen. One of the most common means by which E. coli can be transferred to fresh produce is via contaminated irrigation water. Recognising this potential danger, the World Health Organization (WHO) and the South African Department of Water Affairs (DWA) have set a recommended limit for irrigation water used for fresh produce of 1000 faecal coliforms/100 mL.67
Long-term monitoring studies of the Plankenburg River have revealed that although faecal coliform loads vary depending on the season, the loads are above the recommended limits.8 The Plankenburg River flows past an informal settlement as well as through an industrial area of the town of Stellenbosch before it converges with the Eerste River and flows through an agricultural region, where the water is withdrawn for irrigation. Constant high faecal coliform levels might therefore contribute to the possible transfer of E. coli during irrigation, if river water is not treated prior to irrigation.
Although the presence of several potential pathogens in the Plankenburg River has been reported,8 the occurrence and types of pathogenic E. coli are not known. The aim of this study was therefore to determine the number, types and antibiotic susceptibility of potential pathogenic E. coli present in the Plankenburg River.
Materials and methods
Sampling
Water samples from which E. coli was isolated were collected from the Plankenburg River (Stellenbosch, Western Cape Province) at three sites (P-0, P-1 and P-2).8 The P-0 sampling site was about 5 km upstream of the P-1 site and was selected specifically to assess the impact of an informal settlement and industrial area on the water quality of the Plankenburg River. However, for large parts of each year, especially during the dry summer months, this site had no flowing water and therefore it was not sampled at the same frequency as the other sites. Two of the sites - P-1 and P-2 - are situated downstream of an informal settlement and industrial area and have shown high levels of faecal contamination.8 River water sampling was done in accordance with the SANS 5667-6 method9; all samples were 1 L in volume.
E. coli isolation and identification
Study 1 (year 1):
Daily, for 9 days, 1 L of water was collected aseptically at site P-1. The presence of coliforms, faecal coliforms and E. coli were determined using the multiple tube fermentation (MTF) technique.10 The E. coli broth tubes that exhibited gas production as well as fluorescence after 24 h incubation at 44.5 °C were streaked onto Eosin Methylene-blue Lactose Sucrose (L-EMB) agar (Oxoid, Hampshire, UK). Five typical E. coli colonies (dark purple to black colonies with a distinct metallic sheen) from each of the water samples were selected using Harrison's disc method11 and further purified using Brilliance™ E. col//coliform selective agar (Oxoid).
Study 2 (year 2):
Daily, for 14 days, 1 L of water was collected aseptically at site P-2. E. coli enumeration was done according to the American Public Health Association Standard Method12 and Violet Red Bile Agar (Merck, Johannesburg, South Africa) was used for the enumeration. For further analysis, representative Enterobacteriaceae colonies (red and pink) were selected using Harrison's disc method.11 The selected colonies were further purified using Brilliance™ E. coli/coliform selective agar (Oxoid).
Study 3 (year 3):
As part of a larger environmental study, sites P-0, P-1 and P-2 were sampled twice. Enumeration of total coliforms and E. coli was done according to SANS 930813 using Colilert 18 (IDEXX, Cape Town, South Africa). The Colilert 18 method is considered more user-friendly than the traditional MTF method used in Study 1, and it has been reported that Colilert results compare well with MTF results.14 After incubation,14 Quantitrays were divided into quarters and two fluorescent wells per quarter were pooled (1 mL per well to yield a maximum of 8 mL). A dilution was made from the pooled extract and spread plates were prepared on L-EMB agar (Oxoid). Five typical E. coli colonies from each of the water samples were selected at the highest dilution using Harrison's disc method11 and purified using Brilliance™ E. col/coNform selective agar (Oxoid).
Characterisation and confirmation of E. coli identification
Each isolate was streaked out on nutrient agar (Biolab, Johannesburg, South Africa), and the analytical profile index (API) 20E system (BioMérieux, Johannesburg, South Africa) was used in conjunction with Gram staining, a catalase test and growth on MacConkey medium.15 The profile was then entered into the API Web database (BioMérieux) for species identification. Isolates were stored in 40% (v/v) glycerol at -80 °C.
Polymerase chain reaction analyses
Cell extracts of all isolates were prepared prior to the polymerase chain reaction (PCR) using the method of Altalhi and Hassen16. Isolates identified as E. coli with API were tested for the presence of the E. coli uidA household gene using the primer sequences of Heijnen and Medema17 and KAPATaq™ HotStart DNA polymerase (KAPABiosystems, Cape Town, South Africa). Strains that tested positive for uidA were subjected to a multiplex PCR method as modified from Omar and Barnard18 to screen for the presence of InPEC genes (Table 1). Strains identified as InPECs were also screened for the presence of ExPEC genes19 and classified phylogenetically using the triplex PCR method of Clermont et al.20 for E. coli phylogenetic group (genogroup) identification. The Kapa2G™ Fast Multiplex PCR kit (KAPABiosystems) was used for the InPEC and ExPEC multiplex methods as well as for the triplex PCR method. Positive and negative controls were included in all PCRs, and all reactions were performed in a G-storm thermal cycler (Vacutec, Johannesburg, South Africa). Primer sequences and concentrations are presented in Table 1. Reaction conditions are presented in Table 2. PCR products were all visualised with UV illumination after gel electrophoresis in agarose gels containing 1 ^g/mL ethidium bromide and 0.5 x TBE buffer. Gel electrophoresis was performed at 210V/20 min for the uidA PCR products and the triplex PCR products, using 1.25% agarose gels and 2% agarose gels, respectively. Gel electrophoresis of the InPEC and ExPEC PCR products were conducted at 120V/90 min using 1.25% agarose gels. A 100-bp marker (Promega, Madison, WI, USA) was included for size estimation purposes.
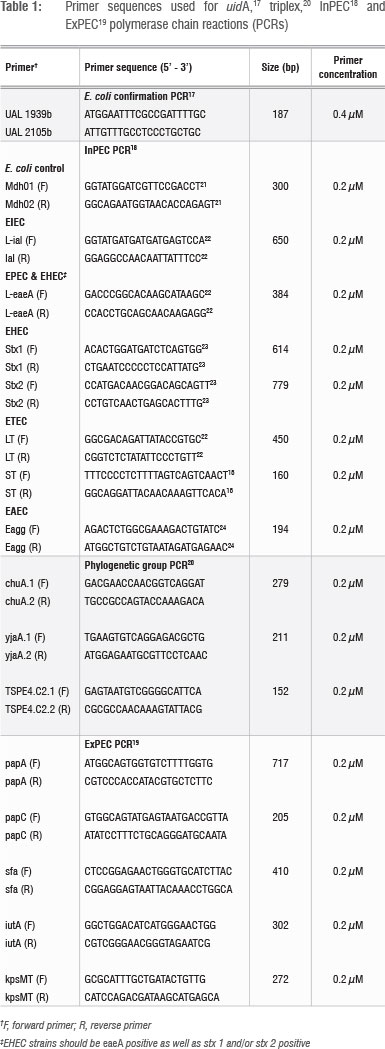
Antimicrobial resistance testing
The E. coli pathogens identified with PCR analysis were also screened for antimicrobial susceptibility against Ampicillin (10 µg), Cephalothin (30 ug), Chloramphenicol (30 ug), Ciprofloxacin (5 µg), Tetracycline (30 µg), Trimethoprim (2.5 µg), and Streptomycin (S) (10 µg). This screening was done using the standard antimicrobial disc susceptibility test described by the US Clinical and Laboratory Standards Institute (CLSI).25 Inhibition zones were interpreted using the performance standards of CLSI26 (for Ampicillin, Cephalothin, Chloramphenicol, Ciprofloxacin, Tetracycline and Streptomycin) and Andrews27 (for Trimethoprim) and strains were classified as susceptible, intermediate or resistant. The American Type Culture Collection (ATCC) strain 25922 was included as a susceptible control in all antimicrobial resistance screening tests.
Standard cultures for PCR analysis
E. coli ATCC 25922 was used as a positive control in the uidA PCR and the triplex PCR. ATCC 25922 was also combined with ATCC 35218 and used as a positive control for the ExPEC multiplex PCR after the identity of bands amplified in the expected regions for papA, sfa/foc, iutA, kpsMT II and papC were confirmed with sequencing and BLAST identification. A combination of EPEC, EHEC, ETEC, EIEC and EAEC positive strains was used as a positive control for the InPEC multiplex PCR.
Results and discussion
E. coli loads
The E. coli loads in the Plankenburg River samples and the number of confirmed E. coli strains are presented in Table 3. In all instances, the E. coli counts exceeded the WHO recommended limit for irrigation water used for fresh produce of 1000 faecal coliforms/100 mL.6 National guidelines set by the DWA associate risk with different E. coli levels in irrigation water: 'no risk' is associated with <1 E. coli/100 mL, 'low risk' is associated with 1-999 E. coli/100 mL and 'high risk' is associated with 1000-3999 E. coli/100 mL.7 Considering these guidelines, the E. coli levels at the time of analysis qualify as unacceptably high for the most part. It would therefore be a fair assumption to conclude that, under these conditions, the transfer of microbes from the Plankenburg River via irrigation onto the surface of fresh produce is highly probable.
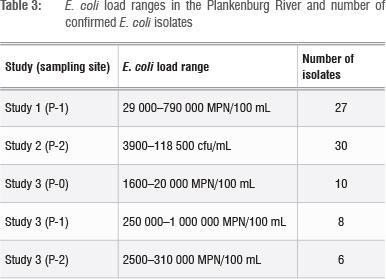
E. coli with InPEC virulence genes
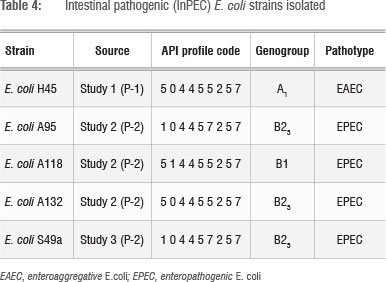
The 81 E. coli strains isolated from the river (Table 3) were positively identified by the API system and their identity was then confirmed using uidA PCR. The strains were then screened for InPEC gene sequences and the characteristics of the pathogenic E. coli strains identified are presented in Table 4. Of the 81 strains screened (Table 3), five pathogens were identified (one EAEC and four EPEC strains) (Table 4). This result concurs with previous reports that E. coli strains that carry pathogenic gene sequences represent 0.9-10% of E. coli present in surface waters.28 It is interesting to note that the three EPEC strains isolated during Study 2 (Table 4) had different biochemical (API) profiles, which suggests that they are not clones.
The API profiles for all the pathogenic strains showed that they differed in three respects: production of lysine decarboxylase (LDC) and L-ornithine decarboxylase (ODC) and utilisation of saccharose. Variation in these three properties has been observed before in E. coli isolated from untreated surface waters and soil.29,30 It has furthermore been reported that E. coli can induce production of amino acid decarboxylases (such as LDC and ODC) in response to reduced pH conditions.31 This report illustrates the highly adaptable nature of E. coli which helps it survive in more acidic environments.32
Because the PCR-based detection method of Clermont20 was used to identify E. coli phylogenetic groups (genogroups), the four main groups (A, B1, B2 and D) can further be subdivided into seven subgroups to increase discrimination: A0, A1, B1, B22, B23, D1 and D2.33 It has been reported that ExPEC strains usually belong to genogroups B2 and D, while InPEC strains that cause severe diarrhoea-related diseases (EIEC, EHEC and ETEC) are most commonly classified into genogroups A and B1.34E. coli strains that cause mild and chronic diarrhoea (EAEC, EPEC and DAEC) can belong to any of the four main phylogenetic groups.34 It has, however, been reported that most human EPEC strains belong to genogroups B1 or B2.35 Carlos et al.36 examined E. coli from a variety of different primary hosts (human, goat, chicken, cow, sheep and pig) and found that B23 strains in particular could only be found in human samples. It was therefore concluded that the EPEC strains isolated from the river in this study were possibly of human origin, which suggests human faecal contamination of the Plankenburg River. The isolation data indicates that the contamination might also be constant over time, as similar EPEC strains were isolated during Studies 2 and 3 (Table 4).
Health implications
The enteroaggregative (EAEC) E. coli, detected during the first study (Table 4), affects the small intestine and causes mild but persistent watery diarrhoea (≥14 days) in people of all ages. It is frequently found in immunocompromised individuals. As a result of mucinase activity, it can cause mild but significant mucosal damage.4,37
Enteropathogenic (EPEC) E. coli, of which four were isolated during Studies 2 and 3 (Table 4), primarily affects children and infants, causing profuse watery diarrhoea, fever and nausea. It can also cause disease in certain animals. Outbreaks in developing countries can be quite severe, with reported mortality rates of up to 30%.37 Adult infections are rare, but possible if high infective doses are combined with substances that can neutralise gastric pH. Certain medical conditions, such as diabetes, can also make adults more susceptible to EPEC infections.38
EPEC attachment to the small intestine is a multi-step process which starts with initial attachment to epithelial cells and microcolony stabilisation through bundle-forming pili (BFP), intimate adherence through the production of intimin, and the formation of attaching and effacing lesions and 'pedestal'-like structures.4,37 Intimin production is facilitated by the locus of enterocyte effacement pathogenicity island which carries the eae gene, and is a prerequisite for EPEC and EHEC pathogenicity. BFP is coded for by the E. coli adherence factor plasmid, and, although it is considered a virulence contributing factor because of its stabilising role during initial microcolony formation, it is not an absolute requirement for EPEC pathogenicity.39 Strains that are eae(+) and BFP(+) are referred to as typical EPEC (tEPEC), while eae(+) and BFP(-) strains are known as atypical EPEC (aEPEC) strains.40 Although symptoms of aEPEC are milder, with non-inflammatory diarrhoea and no fever or vomiting, it is, however, associated with prolonged diarrhoea (>14 days).39 Prolonged diarrhoea for longer than 14 days is clinically associated with an increased risk for illness and death.40 Although the EPEC strains identified in this study were all eae positive, no screening for BFP was done, so the strains could be either tEPEC or aEPEC. However, in both instances, the burden of disease would be significant.
Antibiotic resistance profiles and ExPEC virulence genes
The presence of pathogenic E. coli in this river could result in waterborne or foodborne diseases. Treatment of these diseases could be further complicated if the pathogenic isolates are also resistant to medically important antibiotics. The five InPECs presented in Table 4 were additionally screened for antibiotic susceptibility to seven clinically important antibiotics, as well as for the presence of ExPEC genes. The results are presented in Table 5.
The results showed that the InPEC strains could not be positively confirmed as ExPEC, as they did not have two or more of the ExPEC gene sequences. The EAEC strain (E. coli H45) did carry the ExPEC gene sequence iutA, which codes for the aerobactin siderophore that contributes to essential ferric iron uptake and transport in different iron-deficient environments.41,42 It has been reported that the incidence of aerobactin genes correlates with the presence of highly virulent pathogenic E. coli strains.42 It should, however, be stated that the pathogenic strains were only screened for the most abundant ExPEC gene sequences19 and that more than 30 other ExPEC genes have been reported.41 It is thus possible that E. coli H45 may be a carrier of other ExPEC gene sequences which were not tested for.
The antibiotics tested for in this study all represented different classes of antimicrobials: aminoglycosides (Streptomycin), fluoroquinolones (Ciprofloxacin), penicillins (Ampicillin), amphenicols (Chloramphenicol), cephalosporins (Cefalotin), dihydropholate reductase inhibitors (Trimethoprim) and tetracyclines (Tetracycline). Antibiotic resistance testing revealed that, although all of the InPEC strains were resistant to multiple antibiotics, the four EPEC strains (Table 5) can be referred to as multidrug resistant (MDR) strains. This conclusion is based on the accepted definition of MDR which refers to the co-resistance that a strain can have to three or more classes of antimicrobials.43 This finding concurs with previous observations that multiple antibiotic resistances are common for EPEC.37
The most abundant resistances were against Ampicillin (5/5) and Trimethoprim (4/5), followed by Tetracycline (3/5), Streptomycin (2/5) and Chloramphenicol (1/5). Trimethoprim and Ampicillin resistance were furthermore observed in the InPEC strains from all three studies, which showed that Trimethoprim and Ampicillin resistance persisted among bacteria in the river over time. Tetracycline and Streptomycin resistances were limited to strains from Study 2, while Chloramphenicol resistance was only observed in the isolate from Study 3. Antibiotic resistance can either be carried on mobile genetic elements, such as plasmids, which could be easily transferred horizontally between different bacteria, or be as a result of the environmental selection of a chromosomal mutation.44 Whether chromosomal or plasmid-based, the results showed that antibiotic-resistant E. coli that also carries InPEC virulence genes is present in the Plankenburg River.
E. coli strains resistant to Tetracycline, Streptomycin, Chloramphenicol and Ampicillin have also been isolated from surface and groundwaters in KwaZulu-Natal and the North-West Province.45-47 None of these studies tested for Trimethoprim resistance. The widespread occurrence of resistance to Tetracycline, Streptomycin, Chloramphenicol and Ampicillin is not surprising, as they are 'older' antibiotics, some of which have been in use since the 1940s. The fact that antibiotics such as Tetracycline have also been widely used as growth promoters in animal production, could also have contributed to the extent to which resistance has spread in the environment.48
The increased antibiotic resistance of Gram-negative bacteria such as E. coli is considered a serious global problem, because there is no foreseeable development of new antibiotic classes in the next 10 years.49 This concern has led to the WHO classification of Ampicillin and Streptomycin as antimicrobials of critical importance for human medicine, while Chloramphenicol, Trimethoprim and Tetracycline are considered as highly important antimicrobials.49 The occurrence of E. coli strains that show resistance to these important antibiotics in the Plankenburg River is therefore a matter of concern.
The presence of antibiotic substances in the aquatic environment is increasing mostly as a result of widespread application in the fields of human medical therapy and agriculture. Antibiotic substances are never fully metabolised in humans or other animals, which means that small amounts are regularly excreted and can enter water streams directly or indirectly via faecal pollution, agricultural run-off or through discharge from wastewater treatment plants.50,51 The concentrations are usually at sub-inhibitory levels, which contribute to the development and spread of antibiotic resistance properties among environmental bacteria. If human pathogens acquire resistance to antibiotics, a serious situation can arise which, at the very least, can result in increased disease treatment costs. Antibiotic-resistant pathogens that are resistant to multiple antibiotics might, however, also lead to increased morbidity and mortality rates.51
Conclusions
Escherichia coli counts in the Plankenburg River have been found to be unacceptably high and this river can thus be classified as a 'highrisk' irrigation water source. Under these conditions, transfer of E. coli from the water to produce during irrigation will be highly probable. MDR E. coli strains, harbouring intestinal pathogenic gene sequences, were isolated from the Plankenburg River on more than one occasion. Three of the E. coli strains carrying enteropathogenic sequences also belonged to the B23 phylogenetic group, which could indicate human faecal contamination of the Plankenburg River. Subsequently, if irrigated fresh produce contaminated with MDR pathogens is consumed raw, it might act as a direct vehicle for the transmission of disease. Treatment of these MDR pathogenic bacterial related diseases will then be negatively impacted. It is therefore in the public's interest to report the existence of multiple antibiotic-resistant pathogenic E. coli in the Plankenburg River, so that corrective action, specifically in terms of treatment of irrigation water prior to irrigation, can be taken as soon as possible.
Acknowledgements
We thank Dr T.G. Barnard (Water and Health Research Unit, University of Johannesburg) for kindly providing standard InPEC cultures. This study was part of a solicited research project (K5/1773) funded by the Water Research Commission and co-funded by the Department of Agriculture, Forestry and Fisheries (WRC Knowledge Review 2012_13 KSA4). Financial support provided by the National Research Foundation (IFR2008112100010) is also gratefully acknowledged.
Authors' contributions
T.J.B. and G.O.S. were the project leaders and were responsible for the project design; TJ.B. and C.L. were responsible for the experimental design and supervision; M.R., N.H., A.C., N.S. and C.L. performed the experiments; C.L. and TJ.B. wrote the manuscript; and G.O.S. made editorial contributions.
References
1. Touchon M, Hoede C, Tenaillon O, Barbe V Baeriswyl S, Bidet P et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Gen. 2009;5:e1000344. http://dx.doi.org/10.1371/journal.pgen.1000344 [ Links ]
2. Van Elsas JD, Semenov AV Costa R, Trevors JT. Survival of Escherichia coli in the environment: Fundamental and public health aspects. ISME J. 2011;5:173-183. http://dx.doi.org/10.1038/ismej.2010.80 [ Links ]
3. Liang Z, He Z, Powell CA, Stoffella PJ. Survival of Escherichia coli in soil with modified microbial community composition. Soil Biol Biochem. 2011;43:1591-1599. http://dx.doi.org/10.1016/j.soilbio.2011.04.010 [ Links ]
4. Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nature Rev. 2004;2:123-140. [ Links ]
5. Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg Infec Dis. 2013;19:407-415. http://dx.doi.org/10.3201/eid1903.111866 [ Links ]
6. World Health Organization (WHO). Health guidelines for the use of wastewater in agriculture and aquaculture. Technical report series no. 778. Geneva: WHO; 1989. [ Links ]
7. Department of Water Affairs and Forestry (DWAF). South African water quality guidelines. Volume 4. In: Holmes S, editor. Agricultural use: Irrigation. 2nd ed. Pretoria: CSIR Environmental Services; 1996. p. 63-68. [ Links ]
8. Britz TJ, Sigge GO, Huisamen N, Kikine T, Ackermann A, Loetter M, et al. Fluctuations of indicator and index microbes as indication of pollution over three years in the Plankenburg and Eerste Rivers, Western Cape, South Africa. Water SA. 2013;39:457-465. [ Links ]
9. SANS. Method 5667-6. Water quality - Sampling, Part 6: Guidance on sampling of rivers and streams. Pretoria: Standards South Africa Printers; 2006. [ Links ]
10. Cristensen D, Crawford C, Szabo R. Method MFHPB-19: Enumeration of coliforms, faecal coliforms and of E. coli in foods using the MPN method. Health Products and Food Branch. Ottawa: Food Directorate, Health Canada; 2002. [ Links ]
11. Harrigan W, McCance ME. Statistical methods for the selection and examination of microbial colonies. In: Harrigan W, McCance ME, editors. Laboratory methods in food and dairy microbiology. London: Academic Press; 1976. p. 47-49. [ Links ]
12. American Public Health Association (APHA). Standard methods for the examination of water and wastewater. 21st ed. Washington DC: APHA; 2005. [ Links ]
13. SANS. Method 9308-2. Water quality: Detection and enumeration of coliform organisms, thermotolerant coliform and presumptive Escherichia coli. Part 2: Multiple tube (most probable number) method. Pretoria: Standards South Africa Printers; 2004. [ Links ]
14. Noble RT, Weisberg SB, Leecaster MK, McGee CD, Ritter K, Walker KO, et al. Comparison of beach bacterial water quality indicator measurement methods. Environ Monit Assess. 2003;81:301-312. http://dx.doi.org/10.1023/A:1021397529041 [ Links ]
15. Gerhardt P Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, et al., editors. Manual of methods for general bacteriology. Washington DC: American Society for Microbiology; 1981. [ Links ]
16. Altalhi AD, Hassan SA. Bacterial quality of raw milk investigated by Escherichia coli and isolates analysis for specific virulence-gene markers. Food Cont. 2009;20:913-917. http://dx.doi.org/10.1016/j.foodcont.2009.01.005 [ Links ]
17. Heijnen L, Medema G. Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J Water Health. 2006;4:487-498. [ Links ]
18. Omar KB, Barnard TG. The occurrence of pathogenic Escherichia coli in South African waste water treatment plants as detected by multiplex PCR. Water SA. 2010;36:172-179. [ Links ]
19. Xia X, Meng J, Zhao S, Jones S, Gaines S, Ayers S, et al. Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. J Food Protec. 2011;74:38-44. http://dx.doi.org/10.4315/0362-028X.JFP-10-251 [ Links ]
20. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555-4558. http://dx.doi.org/10.1128/AEM.66.10.4555-4558.2000 [ Links ]
21. Tarr CL, Large TM, Moeller CL, Lacher DW, Tarr PI, Acheson DW, et al. Molecular characterization of a serotype O121 :H19 clone, a distinct shiga toxin-producing clone of pathogenic Escherichia coli. Infec Immun. 2002;70:6853-6859. http://dx.doi.org/10.1128/IAI.70.12.6853-6859.2002 [ Links ]
22. Lopez-Saucedo C, Cerna JF, Villegas-Sepulveda N, Thompson R, Velazquez FR, Torres J, et al. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg Infec Dis. 2003;9:127-131. http://dx.doi.org/10.3201/eid0901.010507 [ Links ]
23. Moses AE, Garbati MA, Egwu GO, Ameh JA. Detection of E. coli O157 and O26 serogroups in human immunodeficiency virus-infected patients with clinical manifestation of diarrhea in Maiduguri, Nigeria. Res J Med Med Sci. 2006;1(4):140-145. [ Links ]
24. Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clinical Microbiol. 2000;38:2001-2004. [ Links ]
25. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial disk susceptibility tests; approved standard. 10th ed. Volume 29 number 1. CLSI document M02-A10. Wayne, PA: CLSI; 2009. [ Links ]
26. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. 31(1). CLSI document M100-S21. Wayne, PA: CLSI; 2011. [ Links ]
27. Andrews JM. BSAC standardized disc susceptibility testing method (version 8). BSAC Working Party on Susceptibility Testing. J Antimicrob Chemother. 2009;64:454-489. http://dx.doi.org/10.1093/jac/dkp244 [ Links ]
28. Masters N, Wiegand A, Ahmed W, Katouli M. Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 2011;45:6321-6333. http://dx.doi.org/10.1016/j.watres.2011.09.018 [ Links ]
29. Janezic KJ, Ferry B, Hendricks EW, Janiga BA, Johnson T, Murphy S, et al. Phenotypic and genotypic characterization of Escherichia coli isolated from untreated surface waters. Open Microbiol J. 2013;7:9-19. http://dx.doi.org/10.2174/1874285801307010009 [ Links ]
30. Brennan FP Abram F, Chinalia FA, Richards KG, O'Flaherty V. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl Environ Microbiol. 2010;76 (7):2175-2180. http://dx.doi.org/10.1128/AEM.01944-09 [ Links ]
31. Kanjee U, Gutsche I, Alexopoulos E, Zhao B, El Bakkouri M, Thibault G, et al. Linkage between the bacterial acid stress and stringent responses: The structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931-944. http://dx.doi.org/10.1038/emboj.2011.5 [ Links ]
32. Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A. 'Black holes' and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943-3948. http://dx.doi.org/10.1073/pnas.957.3943 [ Links ]
33. Escobar-Paramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, et al. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol. 2006;8:1975-1984. http://dx.doi.org/10.1111/j.1462-2920.2006.01077.x [ Links ]
34. Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Fairbrother J, et al. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair river and Detroit river areas. Appl Environ Microbiol. 2007;73:477-484. http://dx.doi.org/10.1128/AEM.01445-06 [ Links ]
35. Tramuta C, Robino P, Nebbia P. Phylogenetic background of attaching and effacing Escherichia coli isolates from animals. Vet Res Commun. 2008;32:433-437. http://dx.doi.org/10.1007/s11259-008-9042-1 [ Links ]
36. Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MIZ, Gomes TAT, et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010;10:161-172. http://dx.doi.org/10.1186/1471-2180-10-161 [ Links ]
37. Torres AG. Intestinal pathogenic Escherichia coli. In: Barrett ADT, Stanberry LR, editors. Vaccines for biodefence and emerging and neglected diseases. New York: Elsevier; 2009. p. 1013-1029. http://dx.doi.org/10.1016/B978-0-12-369408-9.00051-2 [ Links ]
38. Nataro JP Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142-201. [ Links ]
39. Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg Infec Dis. 2006;12:597-603. http://dx.doi.org/10.3201/eid1204.051112 [ Links ]
40. Nataro JP Atypical enteropathogenic Escherichia coli: Typical pathogens? Emerg Infec Dis. 2006;12:696-699. http://dx.doi.org/10.3201/eid1204.060125 [ Links ]
41. Chapman TA, Wu XX Barchia I, Bettelheim KA, Driesen S, Trott D, et al. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol. 2006;72:4782-4795. http://dx.doi.org/10.1128/AEM.02885-05 [ Links ]
42. Garénaux A, Caza M, Dozois CM. The ins and outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet Microb. 2011;153:89-98. http://dx.doi.org/10.1016/j.vetmic.2011.05.023 [ Links ]
43. Doyle MP Loneragan GH, Scott HM, Singer RS. Antimicrobial resistance: Challenges and perspectives (Institute of Food Technologists - Expert Report). Comp Rev Food Sci Food Safety. 2013;12:234-248. http://dx.doi.org/10.1111/1541-4337.12008 [ Links ]
44. Akter S, Rafiq-Un N, Rupa FA, Bari MDL, Hossain MAC. Antibiotic resistance and plasmid profiles in bacteria isolated from market fresh vegetables. Agric Food Anal Bacteriol. 2011;1(2):140-149. [ Links ]
45. Olaniran AO, Naicker K, Pillay B. Antibiotic resistance profiles of Escherichia coli isolates from river sources in Durban, South Africa. Water SA. 2009;25:174-183. [ Links ]
46. Wose Kinge CN, Ateba CN, Kawadza DT. Antibiotic resistance profiles of Escherichia coli isolated from different water sources in the Mmabatho locality North-West Province, South Africa. S Afr J Sci. 2010;106(1/2):44-49. [ Links ]
47. Phokela PT, Ateba CN, Kawadza TK. Assessing antibiotic resistance profiles in Escherichia coli and Salmonella species from groundwater in the Mafikeng area, South Africa. African J Microbiol Res. 2011;5(32):5902-5909. [ Links ]
48. Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, et al. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg Infec Dis. 2012;18(5):741-749. http://dx.doi.org/10.3201/eid1805.111153 [ Links ]
49. World Health Organization (WHO). Critically important antimicrobials for human medicine, 3rd revision. Geneva: WHO; 2012. [ Links ]
50. Lupo A, Coyne S, Berendonk TU. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front Microbiol. 2012;3(18):1-13. [ Links ]
51. Li B, Zhang T. Different removal behaviours of multiple trace antibiotics in municipal wastewater chlorination. Water Res. 2013;47:2970-2982. http://dx.doi.org/10.1016/j.watres.2013.03.001 [ Links ]
 Correspondence:
Correspondence:
Trevor Britz
Department of Food Science
Stellenbosch University
Private Bag X1, Matieland 7600, South Africa
EMAIL: tjbritz@sun.ac.za
Received: 06 Nov. 2013
Revised: 06 Feb. 2014
Accepted: 18 Feb. 2014














