Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.110 n.9-10 Pretoria Oct. 2014
http://dx.doi.org/10.1590/sajs.2014/20130324
RESEARCH ARTICLE
Novel CYP2E1 haplotype identified in a South African cohort
Laura J. Heathfield; Shareefa Dalvie; Kusha Kalideen; Collet Dandara
Division of Human Genetics, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
ABSTRACT
Alcohol abuse accounts for approximately 2.5 million deaths annually and is the third highest risk factor for disease and disability. Alcohol is metabolised by polymorphic enzymes and the status of an individual with respect to alcohol metabolising enzymes may have forensic relevance in post-mortems. Baseline frequencies of gene variants involved in alcohol metabolism need to be established to aid the identification of suitable population-specific polymorphisms to genotype during molecular autopsies. The principal alcohol metabolising enzymes include alcohol dehydrogenase (ADH), aldehyde dehydrogenase (ALDH) and cytochrome P450 2E1 (CYP2E1). Six single nucleotide polymorphisms (SNPs) - rs1229984G>A and rs2066702C>Tin ADH1B, rs671G>A in ALDH2, and rs3813867G>C, rs2031920C>T and rs6413432T>A in CYP2E1 - were genotyped in 150 individuals from four South African populations: Xhosa, Zulu, South African white and South African coloured. Allele frequencies for each SNP in the four population groups were 0-10% for rs1229984A, 2-12% for rs2066702T, 0-2% for rs671A, 1-4% for rs3813867C, 0-1% for rs2031920T and 3-15% for rs6413432A. Haplotype analysis revealed a novel combination of three SNPs in CYP2E1 whose effects on alcohol metabolism need further investigation. Establishment of baseline frequencies adds to our knowledge of genetic variation in alcohol metabolising enzymes and additional research is required to determine the functional significance of this novel CYP2E1 haplotype.
Keywords: CYP2E1; haplotype; alcohol metabolism; molecular autopsy; South Africa.
Introduction
Alcohol abuse accounts for nearly 2.5 million deaths annually and contributes to about 4% of all global deaths.1 On average, 31% of South Africa's population consume alcohol; and the total amount of alcohol consumed per capita is one of the highest in the world.2 Alcohol is the leading abused substance for the majority of South African citizens3 and contributes to more than 60 different types of injuries and diseases1.
Alcohol metabolism can significantly influence drinking behaviour as well as the risk of alcohol dependence.4,5 Ethanol, the main component of alcohol, is primarily converted to acetaldehyde by alcohol dehydrogenases (ADHs). However, ADH enzymes are easily saturated, especially in chronic alcohol consumers, and, in such cases, two additional families of enzymes - cytochrome P450 family 2 subfamily E polypeptide 1 (CYP2E1) and, to a lesser extent, catalase (CAT) - become involved. Acetaldehyde is then converted to acetate via the mitochondrial isoform of aldehyde dehydrogenase (ALDH2).6 Ethanol is eliminated from the body primarily by metabolism (95-98%) with small quantities being excreted via breath (0.7%), sweat (0.1%) and urine (0.3%).7
Acetaldehyde is toxic and must therefore be metabolised as soon as it is formed.6 There is considerable evidence that accumulated acetaldehyde contributes to tissue damage by inducing mitochondrial cell apoptosis. Furthermore, acetaldehyde has been reported to form adducts with dopamine, resulting in the neurotoxin salsolinol, which is thought to contribute towards alcohol dependence.8,9 Under normal circumstances, an estimated 1-2% of acetaldehyde enters the bloodstream,10 but the human body encompasses an efficient defence mechanism in the form of ALDH2, an enzyme with a high affinity for acetaldehyde. However, in cases of binge drinking, an increased concentration of blood acetaldehyde has been reported to induce acute adverse effects such as facial flushing, tachycardia and severe nausea.11 In some individuals, these symptoms lead to the dislike of alcohol,4 while in others, the accumulation of acetaldehyde results in severe illness, ultimately leading to death. Therefore, it is of pharmacogenetics interest to investigate variation in genes coding for alcohol metabolising enzymes - ADH, CYP2E1, CAT and ALDH2. However, CAT was not included in the current study because it is responsible for less than 2% of alcohol metabolism.5
The functions of gene variants involved in alcohol metabolism are fairly well established and their frequencies are reasonably documented in European and Asian populations.11,12 However, this information is lacking in South African populations. Although Warnich et al.13 extensively reviewed allele frequencies of various CYP enzymes in Bantu-speaking South Africans, CYP2E1 was omitted. However, Li et al.14 determined allele frequencies of various CYP2E1 polymorphisms in black male South Africans in relation to oesophageal cancer, and several other novel CYP variants have been observed in some South African populations.14-17 It is thus of interest to ascertain the frequencies of functionally significant gene variants in all South African populations in order to predict their forensic significance in circumstances associated with alcohol metabolism. In this pilot study, we aimed to establish the frequencies of six pharmacogenetically informative single nucleotide polymorphisms (SNPs) in four South African populations.
Methods
Cohort
The cohort consisted of 150 control subjects from four South African population groups: Xhosa (n=34), Zulu (n=40), South African white (n=44) and South African coloured (n=32). Ethnicity of individuals was self-reported and informed consent was obtained from all participants. For the purposes of this study, the South African white group comprised both English-speaking and Afrikaans-speaking white individuals. It should also be noted that some individuals in the South African coloured group exhibit mixed ancestry. The study was carried out according to the Declaration of Helsinki (2008) and was approved by the University of Cape Town's Research Ethics Committee (REC REF: 103/2009).
Genotyping
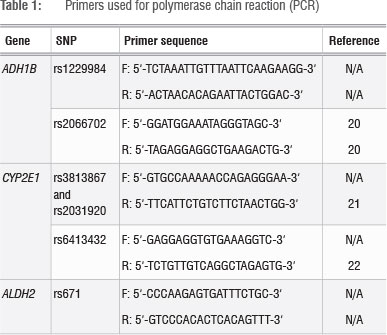
Candidate genes that are involved in alcohol metabolism and their variants were identified using published literature14,18 and data from the 1000 Genomes Project (http://www.1000genomes.org/)19. ADH1B rs1229984G>A (p.Arg49His), ADH1B rs2066702C>T (p.Arg370Cys), CYP2E1 rs3813867G >C, CYP2E1 rs2031920C>T, CYP2E1 rs6413432T>A and ALDH2 rs671G>A (p.Glu504Lys) were selected. Fragments containing the SNPs of interest were amplified using the polymerase chain reaction (PCR). Each PCR mixture contained 100 ng DNA, 5 X Green GoTaq ® reaction buffer (Promega, Madison, WI, USA), 0.5 U Taq polymerase (Promega, Madison, WI, USA), 0.2 mM of each dNTP (Bioline, London, UK) and 0.4 µΜ of each relevant primer (Table 1) in a total volume of 25 µL.20,22 Typical cycling conditions were followed.
SNPs were genotyped using SNaPshot PCR, except for rs6413432 which was genotyped using restriction fragment length polymorphism (RFLP) with the restriction enzyme Dra1. SNaPshot PCR was carried out on cleaned, pooled PCR products, relevant SNaPshot primers (Table 2) and 1 ^L Applied Biosystems SNaPshot® Multiplex Ready Reaction Mix (Applied Biosystems, Carlsbad, CA, USA) in a total volume of 10 ^L. All SNaPshot reactions were carried out on the GeneAmp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA) and cycled 25 times at 96 °C for 10 s, 50 °C for 5 s and 60 °C for 30 s. Samples underwent capillary electrophoresis on the ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA) in the presence of GeneScan™ 120 LIZ® Size Standard (Applied Biosystems, Carlsbad, CA, USA). GeneMapper™ v3.0 (Applied Biosystems, Carlsbad, CA, USA) software was used to analyse peaks and genotypes of samples.
For the RFLP samples were digested with 1 U FastDigest® Dra1 (Fermentas, Ontario, Canada) and 10X FastDigest® Green Buffer (Fermentas, Ontario, Canada) in a final volume of 30 µL and incubated at 37 °C for 1 h. Of the total cohort, 10% was sequenced to verify SNaPshot PCR and RFLP results, whereby at least one representative of each genotype was included for each SNP Each population was represented in this selection.
Statistics
Genotypes for every SNP within each population group were tested for deviation from Hardy-Weinberg equilibrium. Fisher's exact tests were performed to examine whether there were differences in the allelic frequencies between each of the population groups under study, for each SNP independently. The online bioinformatic tool SHEsis (http://analysis.bio-x.cn/SHEsisMain.htm) was used to analyse linkage disequilibrium (LD) between the SNPs under investigation. The SHEsis platform uses a full-precise-iteration algorithm which calculates the probability of the SNPs of interest being inherited together, based on the genotype input data.23,24
Results
All SNPs were in Hardy-Weinberg equilibrium for each population group. Allele frequencies for the six SNPs under investigation are presented in Table 3.19 The South African coloured population exhibited the most heterozygosity for the six SNPs while the Zulu population exhibited the least.
Genotypes for ADH1B rs1229984G>A and rs2066702C>T were analysed in combinations of the previously reported haplotypes ADH1B*1, ADH1B*2 and ADH1B*3 (Table 4).25 All three haplotypes were observed in the Xhosa, South African white and South African coloured populations, but the ADH1B*2 haplotype was absent in the Zulu population. ADH1B*1 was the most frequent haplotype in each population.
Similarly, genotypes for CYP2E1 rs3813867G>C, rs2031920C>T and rs6413432T>A were grouped together into the haplotypes CYP2E1*1A, CYP2E1*5A, CYP2E1*5B and CYP2E1*6 (http://cypalleles.ki.se/cyp2e1.htm) (Table 4). In a total of six cases, CYP2E1 rs3813867 was heterozygous G/C while CYP2E1 rs2031920 and rs6413432 were both homozygous (i.e. rs2031920C/C and rs6413432T/T). This combination of nucleotides (i.e. rs3813867G/C, rs2031920C/C and rs6413432T/T) did not fall into a previously reported haplotype and was noted in a separate row in Table 4.
The CYP2E1*1A haplotype was the most common in all four populations, followed by CYP2E1*6. The CYP2E1*5A haplotype occurred in a single individual in the South African white population while CYP2E1*5B did not appear in any population. The newly observed combination of nucleotides occurred in each of the populations being studied and had the highest relative frequency (0.07) in the Xhosa population.
When comparing allele observations in a pairwise fashion between the four population groups, three significant results were obtained. The Zulu and the South African coloured population groups had significantly different allele frequencies for ADH1B rs1229984A (p=0.009). The ADH1B rs2066702T variant frequency differed significantly between the Xhosa population group and the South African white population group (p=0.034). Allele frequencies for CYP2E1 rs6413432 were significantly different between the Zulu and South African white populations (o=0.030). However, the observed allele frequency distributions were no longer significantly different when the Bonferroni correction for multiple testing was applied.
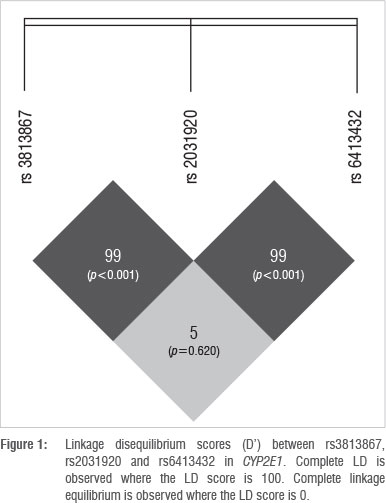
The online bioinformatic tool SHEsis was used to analyse LD between the SNPs in CYP2E1 (Figure 1). The numbers in the blocks within Figure 1 indicate a LD score (D'), whereby an LD score of 100 indicates complete LD. The p-values in brackets indicate the level of significance of the score, calculated using Fisher's exact test.
Discussion
Pharmacogenetics has potential in the clinical setting in which drugs are dispensed for the treatment of disease; however, its use can be extended to aid forensic science, especially in the context of post-mortem in suspected drug-associated deaths. Individuals are frequently exposed to substances which have the capacity to cause death and whose metabolism is influenced by the genetic variants in their metabolising enzyme genes. The utility of pharmacogenetics in forensic science lies in its ability to reduce the miscategorisation of deaths previously classified as sudden unexpected deaths by identification of the associated gene variants that affect metabolism and lead to such deaths. In this study, we aimed to establish the baseline frequencies of variants within genes involved in alcohol metabolism within four South African populations -Xhosa, Zulu, South African white and South African coloured - for future use in predicting possible differences among the population groups in their disposition to alcohol.
Such variants are of forensic relevance as they may help in classifying the manner in which a person might have died. For example, if a postmortem reports a lethal amount of alcohol in the deceased, it may not be readily possible to conclude whether the manner of death was suicidal or accidental26; but the presence of a gene variant associated with accumulation of the particular drug (e.g. ethanol) may point to unintentional accumulation (e.g. of acetaldehyde) and ultimately to accidental death. However, if the individual had a gene variant that resulted in normal metabolism of ethanol, the manner of death would most likely be considered suicide. It has to be kept in mind that substances that inhibit or induce these enzymes are also likely to have similar effects as variant alleles. Therefore, after genotype or phenotype studies, SNPs known to affect metabolism of drugs suspected to have been taken by the deceased, could be recommended for inclusion in a potential molecular autopsy to aid determination of the manner of death.
Of these SNPs with forensic relevance, the South African coloured group exhibited the most heterozygosity and was the only population group to show variation in ALDH2 rs671. However, this finding needs to be confirmed by comparing the distribution of this variant among South African coloured individuals whose deaths were alcohol related and those who drank but whose deaths were not alcohol dependent. Other populations showed relatively little variation in these SNPs; for example, the Zulu population group exhibited variation only in ADH1B rs2066702 (10%), CYP2E1 rs3813867 (1%) and CYP2E1 rs6413432 (3%). With respect to ADH1B, the Zulu group presented with more of the slow ethanol-metabolising variants than the variants associated with fast metabolism.
No individual in the entire cohort contained variants for both ADH1B rs1229984A and ADH1B rs2066702T, which is consistent with the literature to date.20 It can be speculated that either the SNPs are in complete LD or that having both nucleotide changes is incompatible with life.
Although the possibility exists that CYP2E1 haplotypes with different combinations of the three SNPs (rs3813867 (G>C), rs2031920 (C>T) and rs6413432 (T>A)) could occur, current literature does not provide evidence for this. Watanabe et al.27 demonstrated that rs3813867G and rs2031920C are in complete LD. However, a different combination of variants (CYP2E1 rs3813867G/C, CYP2E1 rs2031920C/C) was detected in six individuals in this study, suggesting that a recombination event might have occurred that resulted in the novel haplotype.
One can speculate that if the rs2031920T allele leads to increased transcriptional activation of CYP2E1, and the rs3813867C polymorphism has little effect on transcription,28 individuals with the novel allele detected in this study could more likely display CYP2E1 activity within the normal range. However, functional studies would be needed to confirm this speculation.
The outcome of SHEsis indicated a D' value of 99 for SNPs rs2031920 and rs3813867, indicating that the SNPs are in almost complete LD (p<0.001). The disruption of the former haplotype could be a result of a recombination event and occurred at low frequencies (< 7%) in all four population groups in the study. Non-African populations are known to have larger haplotype blocks as a result of founder effects subsequent to migration out of Africa. Since the out of Africa migration, fewer recombination events have occurred as there have been fewer generations from the new founder group. As a consequence, larger haplotype blocks are inherited by successive offspring,29 which may explain the presence of the novel allele in the Xhosa and Zulu populations. In the South African coloured group, the novel allele could be a result of admixture, resulting in gene flow between black South Africans, the San, the Khoikhoi, western Europeans, Indonesians and Indians who settled in the Cape in the 17th century.30 Therefore, if the recombination event occurred in the African ancestor and the allele was passed to the South African coloured individual in this way, the presence of the novel allele in the South African coloured population is explained. However, this explanation is unlikely as the frequency for the novel allele is second highest for the South African white population group who are of European descent. Rather, a second recombination event could have occurred in the European population group after the out of Africa migration, resulting in the South African coloured group inheriting the allele from the South African white population.
A heritage analysis by Greeff in 2007 revealed that the Afrikaner population also has numerous contributors of genes, including Madagascans, African slaves and Indians,31 which could possibly account for the presence of the haplotype in the South African white population.
Lee et al.32 undertook a comprehensive study and reported global patterns of allele and haplotype frequencies in cYP2E1. They genotyped 11 polymorphisms across cYP2E1 in 50 population groups and showed that allele, haplotype and LD patterns varied greatly among geographical regions. Lee et al.32 identified extensive genetic variation in Africa and reported 16 common haplotypes in addition to various residual haplotypes. An in-depth and direct comparison of haplotype results cannot be made as classification of haplotypes by Lee et al.32 was not done according to the CYP allele database (http://www.cypalleles.ki.se/), as was done here. However, the CYP2E1 SNPs in this study correspond to markers 2 (rs3813867), 3 (rs2070672) and 9 (rs6413432) in Lee et al.'s32 study and fall into 'core groups' A, A and B, respectively. The proposed recombination event in this study was between markers 2 and 3,both of which were grouped into core A by Lee et al.32 It therefore seems unlikely that the novel haplotype proposed in this study has been previously observed, as they both form part of the same core group in this global study.
A limitation of this study was the small sample sizes used for each population group, which may have resulted in an inflation of statistical type 1 and type 2 errors. Furthermore, the self-reported ethnicity of participants also poses a limitation, as baseline frequencies may not have been truly representative of the actual populations. This method was deemed suitable for this pilot study, but a more reliable method to determine ethnicity should be included in subsequent studies.
Baseline frequencies for six informative polymorphisms in genes involved in alcohol metabolism were obtained for four South African populations: Xhosa, Zulu, South African white and South African coloured. The findings reported here add to knowledge in the field, offer a potential utility in the forensic setting and provide a platform for future studies in the area.
Acknowledgements
This work was supported by the University of Cape Town and the National Research Foundation of South Africa.
Authors' contributions
All authors contributed significantly to the study and agreed to the submission of the manuscript. L.J.H. carried out all laboratory experimental work, analysis of results and statistics and wrote the manuscript. S.D., K.K. and C.D. provided conceptual input and were responsible for the project design. C.D. was the project leader.
References
1. World Health Organization (WHO). Global status report on alcohol and health. Geneva: WHO; 2011. [ Links ]
2. Central Drug Authority. Annual report 2007/08. Pretoria: Department of Social Development, Republic of South Africa; 2008. [ Links ]
3. Plüddemann A, Dada S, Bhana A, Perreira T, Carelsen A, Beaufort F, et al. Monitoring alcohol & drug abuse trends in South Africa (July 1996 - June 2008). SECENDU. 2008;11(2):1-12. [ Links ]
4. Hendershot CS, Neighbors C, George WH, McCarthy DM, Wall TL, Liang T, et al. ALDH2, ADH1B and alcohol expectancies: Integrating genetic and learning perspectives. Psychol Addict Behav. 2009;23(3):452-463. http://dx.doi.org/10.1037/a0016629 [ Links ]
5. Agarwal DP Genetic polymorphisms of alcohol metabolizing enzymes. Pathol Biol. 2001;49:703-709. http://dx.doi.org/10.1016/S0369-8114(01)00242-5 [ Links ]
6. Lees GJ. Acetaldehyde: An intermediate in the formation of ethanol from glucose by lactic acid bacteria. J Dairy Res. 1976;43(1):63-73. http://dx.doi.org/10.1017/S0022029900015600 [ Links ]
7. Holford NHG. Clinical pharmacokinetics of ethanol. Clin Pharmacokinet. 1987;13(5):273-292. http://dx.doi.org/10.2165/00003088-198713050-00001 [ Links ]
8. Behrens UJ, Hoerner M, Lasker JM, Lieber CS. Formation of acetaldehyde adducts with ethanol-inducible P450IIE1 in vivo. Biochem Biophysical Res Commun. 1988;154(2):584-590. http://dx.doi.org/10.1016/0006-291X(88)90180-5 [ Links ]
9. Manzo-Avalos S, Saavedra-Molina A. Cellular and mitochondrial effects of alcohol consumption. Int J Environ Res Public Health. 2010;7(12):4281- 4304. http://dx.doi.org/10.3390/ijerph7124281 [ Links ]
10. Nuutinen HU, Salaspuro MP Valle M, Lindros KO. Blood acetaldehyde concentration gradient between hepatic and antecubital venous blood in ethanol-intoxicated alcoholics and controls. Eur J Clin Invest. 1984;14(4):306- 311. http://dx.doi.org/10.1111/j.1365-2362.1984.tb01186.x [ Links ]
11. Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, et al. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: An integrated analysis. Hum Mol Gen. 2008;18(3):580-593. http://dx.doi.org/10.1093/hmg/ddn372 [ Links ]
12. Neafsey P Ginsberg G, Hattis D, Johns DO, Guyton KZ, Sonawane B.Genetic polymorphism in CYP2E1: Population distribution of CYP2E1 activity. J Toxicol Environ Health. 2009;12:362-388. http://dx.doi.org/10.1080/10937400903158359 [ Links ]
13. Warnich L, Drögemöller BI, Pepper MS, Dandara C, Wright GEB. Pharmacogenomic research in South Africa: Lessons learned and future opportunities in the Rainbow Nation. Curr Pharmacogenomics Person Med. 2011;9(3):191-207. http://dx.doi.org/10.2174/187569211796957575 [ Links ]
14. Li D, Dandara C, Parker MI. Association of cytochrome P450 2E1 genetic polymorphisms with squamous cell carcinoma of the oesophagus. Clin Chem Lab Med. 2005;43(4):370-375. http://dx.doi.org/10.1515/CCLM.2005.067 [ Links ]
15. Dodgen TM, Hochfeld WE, Fickl H, Asfaha SM, Durandt C, Rheeder P et al. Introduction of the AmpliChip CYP450 Test to a South African cohort: A platform comparative prospective cohort study. BMC Med Genet. 2013;14:20. http://dx.doi.org/10.1186/1471-2350-14-20 [ Links ]
16. Wright GEB, Niehaus DJH, Drögemöller BI, Koen L, Gaedigk A, Warnich L. Elucidation of CYP2D6 genetic diversity in a unique African population: Implications for the future application of pharmacogenetics in the Xhosa population. Ann Hum Genet. 2010;74(4):340-350. http://dx.doi.org/10.1111/j.1469-1809.2010.00585.x [ Links ]
17. Drögemöller BI, Wright GEB, Niehaus DJH, Koen L, Malan S, Da Silva DM, et al. Characterization of the genetic profile of CYP2C19 in two South African populations. Pharmacogenomics. 2010;11(8):1095-1103. http://dx.doi.org/10.2217/pgs.10.90 [ Links ]
18. Solus JF, Arietta BJ, Harris JR, Sexton DP Steward JQ, McMunn C, et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004;5(7):895-931. http://dx.doi.org/10.1517/14622416.5.7.895 [ Links ]
19. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061-1073. http://dx.doi.org/10.1038/nature09534 [ Links ]
20. Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi A, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;71:84-99. http://dx.doi.org/10.1086/341290 [ Links ]
21. Mittal RD, Srivastava DSL, Mandhani A, Mittal B. Genetic polymorphism of drug metabolizing enzymes (CYP2E1, GSTP1) and susceptibility to bladder cancer in North India. Asian Pac J Cancer Prevent. 2005;6:6-9. [ Links ]
22. Uematsu F, Kikuchi H, Motomiya M, Abe T, Ishioka C, Kanamaru R, et al. Human cytochrome P450IIE1 gene: DraI polymorphism and susceptibility to cancer. Tohoku J Exp Med. 1992;168(2):113-117. http://dx.doi.org/10.1620/tjem.168.113 [ Links ]
23. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97-98. http://dx.doi.org/10.1038/sj.cr.7290272 [ Links ]
24. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19(4):519- 523. http://dx.doi.org/10.1038/cr.2009.33 [ Links ]
25. Osier MV Pakstis AJ, Kidd JR, Lee J-F, Yin S-J, Ko H-C, et al. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet. 1999;64:1147-1157. http://dx.doi.org/10.1086/302317 [ Links ]
26. Sajantila A, Palo JU, Ojanperä I, Davis C, Budowle B. Pharmacogenetics in medico-legal context. Forensic Sci Int. 2010;203:44-52. http://dx.doi.org/10.1016/j.forsciint.2010.09.011 [ Links ]
27. Watanabe J, Hayashi S-I, Nakachi K, Imai K, Suda X Sekine T, et al. Pstl and Rsal RFLPs in complete linkage disequilibrium at the CYP2E1 gene. Nucleic Acids Res. 1990;18(23):7194. http://dx.doi.org/10.1093/nar/18.23.7194 [ Links ]
28. Watanabe J, Hayashi S, Kawajiri K. Different regulation and expression of the human CYP2E1 gene due to the RsaI polymorphism in the 5'-flanking region. J Biochem. 1994;116(2):321-326. [ Links ]
29. Jakobsson M, Scholz SW, Scheet P Gibbs JR, VanLiere JM, Fung H-C, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998-1003. http://dx.doi.org/10.1038/nature06742 [ Links ]
30. De Wit E, Delport W, Rugamika CE, Meintjes A, Möller M, Van Helden PD, et al. Genome-wide analysis of the structure of the South African coloured population in the Western Cape. Hum Genet. 2010;128:145-153. http://dx.doi.org/10.1007/s00439-010-0836-1 [ Links ]
31. Greeff JM. Deconstructing Jaco: Genetic heritage of an Afrikaner. Ann Hum Genet. 2007;71 (5):674-688. http://dx.doi.org/10.1111/j.1469-1809.2007.00363.x [ Links ]
32. Lee M-Y Mukherjee N, Pakstis AJ, Khaliq S, Mohyuddin A, Mehdi SQ, et al. Global patterns of variation in allele and haplotype frequencies and linkage disequilibrium across the CYP2E1 gene. Pharmacogenomics J. 2008;8(5):349-356. http://dx.doi.org/10.1038/tpj.2008.9 [ Links ]
 Correspondence:
Correspondence:
Laura Heathfield
Division of Human Genetics, Institute of Infectious Disease and Molecular Medicine
Faculty of Health Sciences, University of Cape Town
Anzio Road, Observatory 7925, South Africa
EMAIL: laurajaneheathfield@gmail.com
Received: 11 Oct. 2013
Revised: 07 Feb. 2014
Accepted: 18 Feb. 2014














