Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.109 n.11-12 Pretoria Jan. 2013
RESEARCH ARTICLE
Expression of defence-related genes against Phytophthora cinnamomi in five avocado rootstocks
Juanita EngelbrechtI; Noëlani van den BergII
IDepartment of Microbiology and Plant Pathology, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
IIDepartment of Genetics, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
ABSTRACT
Avocado (Persea americana) - a major fruit crop worldwide - is threatened by root rot caused by Phytophthora cinnamomi. This pathogen is known to infect the plant via the feeder roots leading to branch dieback, and eventually tree mortality. While it is known that different avocado rootstocks have varying degrees of susceptibility to Phytophthora root rot, little research has been done on the avocado-Phytophthora interaction. In this study, transcript abundance levels of defence-related genes coding for phenylalanine ammonia-lyase, lipoxygenase, pathogenesis-related protein 5, endochitinase, gluthathionine S-transferase and metallothionein were characterised and compared among five rootstocks with varying susceptibility to root rot, after exposure to P cinnamomi. Root samples were collected at 0 h, 3 h, 6 h, 12 h, 24 h, 48 h and 72 h post-infection and transcript abundance of the defence-related genes was determined using quantitative real-time reverse transcription PCR. The results indicated the involvement of PR-5 and endochitinase in the defence response of all avocado rootstocks to P cinnamomi but these genes could not be directly linked to the observed phenotypic resistance. PR-5 and endochitinase were highly upregulated at 72 h post-infection. Differences in transcript abundance of phenylalanine ammonia-lyase and lipoxygenase genes were seen when comparing tolerant and less tolerant rootstocks, which may suggest that transcripts of these genes contribute to resistance. These data provide important insights into plant defence and into how different avocado rootstocks may exhibit increased resistance to infection by P. cinnamomi.
Keywords: Phytophthora cinnamomi; gene expression; resistance; avocado; defence
Introduction
Avocados (Persea Americana Mill.) are susceptible to a wide range of pathogens and pests and it is therefore not surprising that Phytophthora root rot (PRR) is one of the most damaging diseases, and severely affects commercial production worldwide. The global destructiveness caused by Phytophthora cinnamomi Rands has made it one of the most economically important groups of plant pathogens in the world and also one of the most studied. Primary infection by P. cinnamomi occurs at the small absorbing avocado feeder roots, resulting in a brownish black and brittle appearance. There is almost no progression into the larger roots.1 This root rot leads to death of the feeder roots which results in insufficient water and nutrient uptake, which eventually leads to tree mortality.2
Although PRR has been studied for more than 60 years, total control remains elusive and economic losses continue to increase. Currently, control is achieved by spraying or injecting trees with phosphite in conjunction with using tolerant rootstocks. However, in 2008, Dobrowolski et al.3 showed a decreased pathogen sensitivity to phosphite in Australian avocado orchards where phosphite had been used against P. cinnamomi for prolonged periods.3 Although this decreased sensitivity is currently not viewed as a major threat, the potential impact of such a phenomenon on the effectiveness of phosphite for PRR control may be severely negative. In addition to chemical control, much emphasis is currently placed on the selection and use of tolerant avocado rootstocks.4
Tolerant rootstocks offer the greatest possibility of a sustainable long-term solution for root rot. Several breeding and selection programmes around the world have identified rootstocks with a high degree of resistance to P. cinnamomi5 'Duke 7' was discovered by Zentmyer and in 1975 it became the first commercial rootstock with moderate resistance against the pathogen. It has been a highly successful rootstock and is still used worldwide, although several newer varieties have been selected since then.1 The fact that Duke 7 can acclimatise to many different environmental conditions makes it a popular rootstock today. Other tolerant varieties include 'Thomas', 'Toro Canyon', 'Martin Grande', 'Spencer' and 'G755'. 'Dusa®' ('Merensky 2'), a new variety selected by Westfalia Technological Services, has surpassed the performance of Duke 7, especially under South African climatic conditions.6
Although defence responses have been vigorously studied in model plant species, there is a vast amount yet to be discovered in non-model plants in order to comprehend their underlying defence mechanisms. Moreover, defence mechanisms can vary between different plant species and therefore each particular pathosystem of interest should be studied individually. Numerous studies have shown that salicylic acid is more involved in defence responses associated with biotrophic pathogens whereas jasmonic acid and ethylene are more involved against necrotrophic pathogens and insects.7,8 Therefore, depending on the lifestyle of a pathogen, different defence mechanisms will be activated. Phytophthora cinnamomi is a hemibiotroph - that is, it has both biotrophic and necrotrophic phases9,10 -which is a great advantage because it is capable of switching to a necrotrophic phase when plant mechanisms are encountered that limit its spread. In such a case, a plant defence mechanism such as the hypersensitive response would be ineffective against P. cinnamomi because the pathogen switches to the necrotrophic phase and uses the dead tissue as a nutrient source.
Traditionally, genetic resistance has been classified into two types: qualitative and quantitative resistance. Qualitative resistance is mediated by R-genes and only provides short-lived resistance in the field as new virulent races of the pathogen rapidly overcome resistance encoded by single resistance genes. In contrast, quantitative resistance is controlled by multiple interacting genes that do not prevent infection, but slow down the development of the pathogen and, hence, lasts longer. In the case of P. cinnamomi and avocado, R-mediated resistance is unlikely to be the mechanism responsible for conferring resistance. A multigenic trait is more likely as various levels of resistance in avocado rootstocks against P. cinnamomi have been documented.
When P. cinnamomi infects avocado various defence responses are induced, but few molecular studies have elucidated this interaction. In studies by Garcia-Pineda et al.11 on the defence response in avocado roots against P. cinnamomi, a reactive oxygen species burst as well as the involvement of catalase, epicathecin and nitric oxide were also highlighted. Müniz et al.12 used a proteomics approach to report on protein induction in response to infection of P cinnamomi in roots of avocado. Proteins that were found to be involved in this interaction included isoflavone reductase, glutathione S-transferase, several abscisic acid-stress-ripening proteins, cinnamyl alcohol dehydrogenase, cinnamyl-CoA reductase, cysteine synthase and quinone reductase. These studies have barely begun to unravel how defence against P. cinnamomi is mediated within avocado and a comprehensive analysis of differentially expressed genes could contribute to a better understanding of the molecular processes involved in conferring resistance to PRR. Knowledge of the genetic basis of this observed resistance would not only contribute to the understanding of defence mechanisms but also aid in the development of superior avocado rootstocks.
Therefore, in this study, we elucidated the transcript abundance level of known plant defence-related genes in the response against P cinnamomi in five avocado rootstocks that vary in resistance to PRR. The transcript abundance of these genes was investigated using the quantitative reverse transcription polymerase chain reaction (qRT-PCR) over a time course following inoculation with P. cinnamomi.
Materials and methods
Plant material
A hydroponic system previously used for banana13 was adapted for the avocado-P. cinnamomi pathosystem. Five avocado rootstocks were selected based on their resistance level to P. cinnamomi. These rootstocks were: R0.06 (highly tolerant), Dusa® (highly tolerant), Duke 7 (tolerant), R0.01 (tolerant) and R0.12 (least tolerant); the rootstocks were obtained from Westfalia Technological Services (Tzaneen, Limpopo, South Africa). The 9-month-old clonally propagated plantlets were removed from their bags and transplanted into 500-mL polystyrene cups filled with water to ensure easy access to the roots.
Inoculum preparation
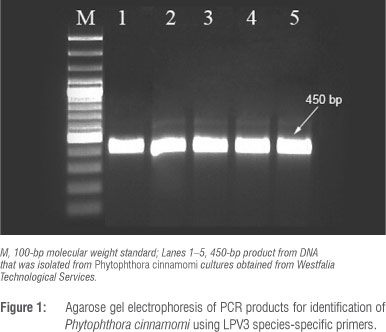
A P. cinnamomi isolate provided by Westfalia Technological Services was used as the inoculum source. Prior to inoculation, the identity of this isolate was confirmed by amplifying the species-specific LPV3 fragment using LPV3 F (5' GTGCAGACTGTCGATGTG 3') and LPV3 R (5' GAACCACAACAGGCACGT 3') primers.14 Genomic DNA was extracted from mycelia growing on ½ PDA (10 g potato dextrose, 15 g agar) using PrepMan™ Ultra Reagent (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer's instructions. The 20-µL PCR reaction contained 2.5 µL 10x PCR reaction buffer, 2.5 mM MgCl2, 200 µM dNTP, 0.25 of each specific primer, 1 U Taq polymerase (Bioline Ltd, London, UK) and 20-50 ng of template DNA. PCR cycling conditions were: an initial denaturation step at 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 58.5 °C for 30 s and 72 °C for 1 min and a final extension step at 72 °C for 7 min. PCR products were separated and examined by electrophoresis on 2% agarose with GelRed (Biotium Inc., Hayward, CA, USA) and visualised under UV light.
Mycelial suspension
The P. cinnamomi isolate was grown on ½ PDA at 25 °C for 4 days, whereafter a sterile broth (8 g D-glucose, 0.8 g yeast extract, 800 mL distilled water, autoclaved at 121 °C for 15 min) was inoculated with six agar blocks (10 mm x 5 mm) containing mycelia and shaken at 25 °C at 150 rpm for 10-14 days. Mycelial balls were placed on Whatman filter paper and left to dry briefly. Mycelia (3.05 g) were mixed with 1 L sterile water and blended for a few minutes to macerate the mycelial pieces. The solution was mixed with 5 L of sterile water and used for inoculation.
Zoospore suspension
Phytophthora cinnamomi was first grown on V8 agar plates (50 mL of filtrated V8 juice, 0.5 g CaCO3, 20 g agar) for 5 days. Small agar blocks (10 mm x 5 mm) containing mycelia were cut at the actively growing margin of the plates and transferred onto empty plates, to which 25 mL of 2% V8 broth (20 mL V8 juice with 0.5 g CaCO3) was added and left for at least 3 days at room temperature (ca. 25 °C). The broth was removed and agar blocks containing mycelia were rinsed three times with distilled water after which 25 mL filtered stream water was added to each of the plates and incubated for 2-3 days at room temperature under UV light. Sporangia formation was monitored during this incubation step by using a Zeiss Stemi 2000 stereomicroscope (Carl Zeiss Ltd., Munich, Germany). Once sufficient sporangia formation was observed, the cultures were cold shocked by placing them at 4 °C for 45 min after which they were removed immediately and left at room temperature for 1 h to stimulate the release of zoospores. The zoospore suspension was decanted from the plates, pooled together and used for inoculation.
Inoculation and sample collection
Avocado roots were suspended in 500-mL polystyrene cups and inoculated with a mixture containing 50 mL macerated mycelia and 1.5 x 103/mL zoospores. Control plants received sterile water instead of inoculum. Root tissue for RNA extraction was collected at 0 h, 3 h, 6 h, 12 h, 24 h, 48 h and 72 h post-infection (hpi). Because of a limited number of avocado plants, we did not include uninfected controls at each time point for this study. Root material from three to five plants per avocado rootstock was harvested per time point to form three biological samples representing either one or two avocado plants pooled together. Root tissue was immediately frozen in liquid nitrogen, ground to a fine powder with a homogeniser (IKA A11 United Scientific (Pty) Ltd, San Diego, CA, USA) and stored at -80 °C. After inoculation, plants were evaluated for root rot symptoms. Selected infected plants were transplanted to plastic bags containing perlite and kept for 6 weeks to confirm the development of PRR symptoms.
Gene expression profiling
RNA extraction and cDNA synthesis
RNA was extracted from root powder using a modification of the CTAB RNA extraction method developed by Chang et al.15 and stored at -80 °C. It is known that avocados have a high polysaccharides content, which seems to influence the quality of RNA; therefore the number of chloroform extractions and centrifugation steps was increased to four. Concentration of RNA was determined using a Nanodrop ND-100 Spectrophotometer (Nanodrop Technologies Inc., Montchanin, DE, USA). RNA integrity was assessed under non-denaturing conditions as described. DNase treatment of RNA was performed by the addition of 1 U RNase-free DNase (Fermentas Life Sciences, Hanover, MD, USA), 1 µL 10x reaction buffer with MgCl2, 1 µg RNA and diethylpyrocarbonate-treated water to a final volume of 9 µL. The mixture was incubated at 37 °C for 30 min followed by the addition of 25 mM EDTA and incubation at 65 °C for 10 min. DNase-treated RNA was column purified using the RNeasy® MiniEluteTM Cleanup kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
First-strand cDNA synthesis was carried out in a total volume of 5 µL which consisted of 0.5 random hexamers (Invitrogen Life Sciences, CA, USA), 1 µg RNA from the previous step and RNase free water. The mixture was incubated at 70 °C for 5 min and then chilled on ice for 5 min, followed by the addition of 40 U RNase inhibitor (Fermentas, Ontario, Canada), 0.5 mM dNTPs (Fermentas, Ontario, Canada), 3 mM MgCl2, 4 µL 5x ImProm-II™ reaction buffer and 1 µL ImProm-II™ reverse transcriptase (Promega Corporation, Madison, WI, USA). Finally the mixture was incubated at 25 °C for 10 min followed by 60 min at 42 °C and 10 min at 70 °C.
The cDNA was analysed for genomic DNA contamination by PCR using the gene-specific primers F3H-F (5' TCTGATTTCGGAGATGACTCGC 3') and F3H-R (5' TGTAGACTTGGGCCACCTCTTT 3') which flank an intron of the flavanone 3-hydroxylase (F3H) gene. PCR amplifications were carried out using first-strand cDNA as the template. The PCR reaction mixture of 20 µL final volume contained 2.5 µL 10x PCR reaction buffer, 2.5 mM MgCl2, 200 µM dNTP 0.25 µM of each specific primer, 1 U Taq polymerase and 1 µl cDNA and water. Amplifications were performed in an Eppendorf MasterCycler gradient (Eppendorf, Hamburg, Germany) under the following conditions: an initial denaturation at 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min. Final extension was carried out at 72 °C for 7 min. PCR products were visualised as before.
Real-time RT-PCR primer design
The transcript abundance of six avocado defence-related genes -phenylalanine ammonia-lyase (PAL), lipoxygenase (LOX), pathogenesis-related protein 5 (PR-5), endochitinase, gluthathionine S-transferase (GTH) and metallothionein - was investigated. Actin and 18S rRNA were used as endogenous controls. Primers for these genes were designed from sequences obtained from the NCBI database by using Primer 3 software and synthesised by either Operon Biotechnologies GmbH (Cologne, Germany) or Inqaba Biotec (Pretoria, South Africa) (Table 1).
Primer pairs were tested for successful amplification of target genes with cDNA in a conventional PCR assay. The PCR reactions were carried out in a total volume of 20 µL containing 2.5 µL 10x PCR reaction buffer, 2.5 mM MgCl2, 200 µM dNTP 0.25 µM of each specific primer, 1 U Taq polymerase and 1 µL of cDNA. The cycling conditions were 95 °C for 3 min, followed by 44 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, with a final elongation step at 72 °C for 10 min. PCR products were analysed as before.
Gene expression analysis using real-time qRT-PCR
A dilution series was performed and a standard curve was generated for each gene to examine the linearity of amplification over a dynamic range. Quantitative RT-PCR was performed using the Bio-Rad® CFX 96 instrument (Bio-Rad, Hercules, CA, USA). Serial dilutions (1:10, 1:25, 1:50, 1:100 and 1:1000) were performed using 5 µL of diluted cDNA from a mixture of all treatment samples inoculated with the pathogen to calculate the standard regression curves. Each dilution was performed in triplicate. Control treatments contained water as template. All PCR reactions were performed in triplicate on each of the three independent biological replications. A 20-µL reaction for PCR amplification contained 10 µL SensimixTM SYBR No-ROX (Bioline Ltd, London, UK), 1 µL of each of the forward and reverse primers (10 µM), 5 µL diluted (1:5) cDNA template and 3 µL PCR-grade water (Roche Applied Science, Mannheim, Germany). The cycling conditions were as follows: pre-incubation for 10 min at 95 °C (hot start) followed by 44 cycles, each consisting of 10 s denaturing at 95 °C, 10 s annealing at 59 °C, 10 s primer extension at 72 °C and data acquisition at 95 °C. Melting curve analysis of the qRT-PCR products was performed to confirm that the individual qRT-PCR signals corresponded to a single homogenous cDNA product.
Data analyses
Standard regression curves were calculated from amplification data of the serial dilutions as follows: y=mx+b, where b is the y-intercept of the standard curve line (crossing point) and m is the slope of the standard curve line (a function of PCR efficiency).16 The resulting crossing point values for each input amount of template were plotted as a function of the log10 concentration of input amounts and a linear trend line was imposed on the data. R2 is the proportion of variability that the data set is accounted for by a statistical model; thus, R2=1 indicates that the fitted model explains all variability in y, while R2=0 indicates no linear relationship between the response variable and regressors. Statistical significance from the qRT-PCR data was analysed by one-way analysis of variance followed by a Student's multiple comparison t-test using the software package JMP 9 (SAS Institute Inc., Cary, NC, USA). In all cases significance was evaluated at p<0.05.
Results
Confirmation of P cinnamomi and symptom evaluation
The identity of the P. cinnamomi isolate was confirmed. The amplification of a region of the LPV3 gene from DNA yielded the expected amplicon size of 450 bp (Figure 1). Avocado plants (five rootstocks) infected with P. cinnamomi developed typical root rot symptoms 6 weeks after infection. Differences between rootstocks were clearly visible 2 weeks after infection (Figure 2). These symptoms included necrotic lesions that developed on the roots and lower stem and some of the feeder roots were black and brittle. Plants showed yellowing and wilting of leaves and, in severe cases, leaf abscission and death. The highly tolerant R0.06 appeared the healthiest when compared to the least tolerant rootstock (R0.12) which showed severe symptoms. The pathogen was re-isolated and Koch's postulates were proven.
RNA extraction and cDNA synthesis
Avocado root RNA samples were of high quality and the absence of contaminating genomic DNA was confirmed for all cDNA samples. The amplification of a region of the F3H gene from cDNA yielded the expected 241-bp RNA-derived product, which was clearly distinguishable from the 1221-bp genomic DNA-derived, intron-containing product (Supplementary figure 1 online).
Real-time RT-PCR primer design
Actin and 18S rRNA consistently exhibited M values (reference gene stability values) less than 0.50 (0.1543 and 0.2508, respectively) and therefore were selected as endogenous controls for this study. 'Gene stability is expressed as an M value, which is inversely proportional to the variation in expression for a given gene.'17 The lower the M-value, the more stably expressed the reference gene is.
All primer pairs designed for defence-related genes PAL, LOX, PR-5, endochitinase, GTH and metallothionein produced single products between 75 bp and 150 bp as expected. The optimal annealing temperature for all the primer pairs was 59 °C (Table 1). The standard curves provided insight into the PCR efficiency for each particular primer set (Supplementary figure 2 online).
Gene expression profiling
Basal (0 hpi) and early transcript levels (3 hpi, 6 hpi and 12 hpi) of PR-5 were very low and similar in tolerant and less tolerant avocado rootstocks (Figure 3a). The first significant response after P. cinnamomi infection occurred at 24 hpi in the moderately tolerant Duke 7. By 48 hpi all rootstocks (except Duke 7) had a significant increase in PR-5. The least tolerant R0.12 and highly tolerant R0.06 had the highest transcript abundance at 48 hpi. By 72 hpi PR-5 transcript levels in all rootstocks continued to increase significantly. The highest transcript abundance of PR-5 was observed at 72 hpi for all rootstocks. R0.06 and Dusa had a 42- and 24-fold increase, respectively, compared with uninfected roots (0 hpi); the least tolerant rootstock (R0.12) also showed a 19-fold increase (Figure 3a).
Basal transcript levels of PAL were significantly higher in the least tolerant R0.12 compared with all rootstocks except Dusa (Figure 3b). By 3 hpi, all rootstocks (except R0.01) showed a decrease in PAL levels (although not significant). A significant increase in PAL was observed in the highly tolerant Dusa at 6 hpi. PAL levels in the less tolerant R0.12 and moderately tolerant Duke 7 continued to decrease significantly at 6 hpi whereas R0.06 and Dusa only decreased at 12 hpi. By 24 hpi, R0.06 and Dusa once again had increased significantly but levels of PAL between the different rootstocks were similar (Figure 3b). At 48 hpi, PAL transcript levels in R0.12 and Duke 7 were significantly lower than in the other rootstocks and remained low at 72 hpi. The levels in all other rootstocks remained similar at 48 hpi when compared with levels at 24 hpi. By 72 hpi, the highly tolerant Dusa also exhibited a significant decrease in PAL levels (Figure 3b). Noteworthy is that transcript levels of PAL in the less tolerant R0.12 continued to decrease over the entire time course.
The constitutive abundance of LOX transcripts was significantly higher in the highly tolerant Dusa when compared with the other rootstocks. LOX levels in Dusa had decreased significantly by 3 hpi (Figure 3c). The levels remained similar in all rootstocks at 6 hpi and 12 hpi. By 24 hpi, Duke 7 and Dusa showed significantly higher levels when compared with the same rootstocks at 12 hpi and when compared with other rootstocks at 24 hpi. LOX levels in R0.01 and less tolerant R0.12 increased significantly from 24 hpi to 48 hpi. At 72 hpi, R0.06 and R0.01 showed a significant increase in LOX, with transcript levels in R0.01 being the highest across all time points and rootstocks (Figure 3c). R0.01 showed an 8-fold increase at 72 hpi compared with 0 hpi.
Basal levels of endochitinase transcripts were low in all rootstocks and significantly lower in highly tolerant R0.06 and moderately tolerant R0.01 when compared with less tolerant R0.12. Levels remained similar at 3 hpi, 6 hpi and 12 hpi. The industry standard (Duke 7) exhibited a significant increase at 12 hpi (Figure 3d). The highly tolerant rootstocks R0.06 and Dusa as well as R0.01 showed significant increases in endochitinase transcript levels at 24 hpi, compared with the less tolerant R0.12 that remained unchanged from 12 hpi (Figure 3d). By 48 hpi, levels of endochitinase continued to increase in R0.06 and Dusa. Highly tolerant Dusa had the highest transcript abundance of endochitinase at 48 hpi; by this time R0.12, Duke 7 and R0.01 also showed significant increases in endochitinase (Figure 3d). At 72 hpi, endochitinase transcripts in R0.06, R0.01 and Duke 7 continued to increase compared with a decrease in the less tolerant R0.12, whereas endochitinase transcript levels in Dusa remained high. Endochitinase transcript abundance levels in R0.01 were significantly the highest at 72 hpi (Figure 3d).
Metallothionein transcript levels were significantly higher in the highly tolerant Dusa when compared with the less tolerant R0.12 and Duke 7 before infection with P. cinamomi. The only significant response after pathogen infection was an increase in metallothionein in the less tolerant R0.12 at 3 hpi, followed by a sharp decrease at 6 hpi (Figure 3e). R0.06 and Dusa had significantly increased transcript levels of metallothionein compared with less tolerant R0.12 at 6 hpi and 12 hpi, respectively. At 12 hpi, Duke 7 also exhibited a significant increase in metallothionein transcripts. Noteworthy is the constant high level of metallothionein in Dusa over the first 24 h. By 24 hpi, the highly tolerant R0.06 had a transcript abundance level of 1.2, resulting in a significantly higher level compared with less tolerant R0.12 (Figure 3e). By 48 hpi, all rootstocks, except the highly tolerant R0.06 and Dusa, showed significant decreases in metallothionein. Furthermore, R0.06 and Dusa continued to have higher transcript levels of metallothionein compared to less tolerant R0.12. By 72 hpi, metallothionein transcript levels in all rootstocks had declined; however, the highly tolerant R0.06 remained significantly higher compared with all other rootstocks (Figure 3e).
Basal transcript abundance of GTH did not differ significantly among R0.12, Duke 7, R0.06 and Dusa. Levels remained unchanged by 3 hpi and 6 hpi; except in the case of Duke 7 which showed a significant decrease at 6 hpi (Figure 3f). By 12 hpi, levels of GTH in Duke 7 had increased significantly. At 24 hpi, the less tolerant R0.12 and tolerant R0.01 exhibited significant increases in GTH levels. By 48 hpi, all rootstocks, except highly tolerant R0.06 and Dusa, displayed significant decreases in GTH. At 72 hpi, the less tolerant R0.12 had the lowest GTH of all rootstocks (Figure 3f).
Discussion
This is the first study of its kind aimed at unravelling the regulation of a set of six defence-associated genes during the first 72 hpi in five avocado rootstocks with different levels of resistance/susceptibility to PRR. A comprehensive understanding of the molecular mechanisms involved during the tolerant host response will enable the identification of defence marker genes that in turn will aid in the selection of avocado rootstocks with enhanced PRR resistance. Phenotypic PRR resistance data for R0.06 and Dusa has been collected over at least a decade by Westfalia Technological Services and both these rootstocks have consistently shown high levels of tolerance in the field under various climatic conditions. In addition to PRR tolerance, both rootstocks have produced high yields when grafted with Hass. In South Africa, 'Dusa®' has replaced Duke 7 as the preferred rootstock for commercial avocado production. Rootstock R0.12 has the lowest level of resistance of all the rootstocks examined in this study. However, it possesses some tolerant attributes and was initially selected in greenhouse trials as a promising candidate but failed during field trials. While there were no uninfected control plants for each time point, because of a limited number of plants, the data generated from this study still provides insights into the role of the selected genes in conferring resistance to PRR. Our results provide a deeper understanding of the phenotypic resistance observed in a previous study on the Phytophthora-avocado interaction, in which the degree of resistance was monitored by qPCR whereby Phytophthora DNA was quantified in Dusa and R0.12. The level of P. cinnamomi in roots of R0.12 was significantly higher when compared with Dusa plants at all time points investigated.18
PR-5 was significantly upregulated at 24 hpi, 48 hpi and 72 hpi in all rootstock varieties infected with P. cinnamomi. The slow but continuous upregulation of PR-5 in all avocado rootstocks suggests that the gene may be part of an important early defence strategy against the biotrophic P. cinnamomi. PR proteins are defined as localised proteins (intra- and extracellular) that accumulate in distant plant tissue after pathogen attack.19 For many years PR proteins have been correlated with the development of systemic acquired resistance.20 Although they are widely used as markers for systemic acquired resistance, their exact roles have not yet been identified. It has been shown that the induction of PR-5 requires salicylic acid signalling and that salicylic acid signalling is more associated with defence against biotrophs.7,8 The fact that PR-5 was also upregulated in the less tolerant rootstock (R0.12) was expected as previous studies have shown that R0.12 possessed resistance against P. cinnamomi, albeit weak. From this study it seems that PR-5 transcript abundance levels cannot be directly correlated with phenotypic PRR resistance, but the gene is significantly induced in response to P. cinnamomi infection.
Expression analysis of PAL transcripts after infection with P. cinnamomi showed that the less tolerant R0.12 and Duke 7 rootstocks showed strong downregulation of PAL transcripts. It can be hypothesised that this downregulation of PAL might aid establishment and development of P. cinnamomi in the root tissue. Upon recognition of P. cinnamomi by avocado, a series of signalling pathways is activated. Among these, the phenylpropanoid pathway plays an extremely important role in secondary plant metabolism by producing numerous phenolic propanoids such as phenolic acids and flavonoids that have vital structural- and defence-related functions.21,22 Phenylalanine ammonialyase is one of the fundamental enzymes in the phenylpropanoid pathway. Therefore it is possible that more phenolic compounds are produced in the highly tolerant rootstocks to combat P. cinnamomi growth whereas the downregulation in the less tolerant rootstocks implies that fewer phenolic compounds are being produced. In pepper (Capsicum annuum L.), an increase in phenolic acids reduced lesion length and invasion upon infection by Phytophthora capsici, whereas susceptible cultivars of C. annuum produced lower amounts of phenolic acids and lesion lengths were not reduced.23
Inoculation of avocado plants resulted in significant increased levels of LOX transcript levels in Dusa and Duke 7 at 24 hpi. An increase in LOX has also been reported to occur in other plant-pathogen interactions.24-26 Lipoxygenases (EC 1.13.1 1.12) are a family of enzymes that catalyse the dioxygenation of polyunsaturated fatty acids in lipids. The role of LOX in defence against pathogens is connected to the production of compounds that are involved in signalling27, antimicrobial activity28, and hypersensitive response development29. The fact that R0.12 remained unchanged during the first 24 h indicates a delayed response and therefore subsequent failure to restrict the pathogen by not allowing signalling of other defence responses or by a lack of antimicrobial activity. Tomato plants infected with Phytophthora infestans have been shown to display fungitoxic activities by producing linolenic acid via lipoxygenase.30
An increase in transcript levels of endochitinase became evident at 24 hpi in all rootstocks with the exception of R0.12. R0.12 was the only rootstock that did not show any significant changes during the first 24 hpi, which suggests that the delay of 24 h allowed establishment of P. cinnamomi in the less tolerant rootstock when compared with other rootstocks that showed significant upregulation at 24 hpi. Endochitinases play a role in plant defence by attacking structural chitin present in the cell wall of fungi.31 Unlike fungal cell walls, oomycete cell walls are mainly composed of cellulosic compounds and glucans but have been found to contain limited amounts of chitin.32,33 Consequently, limited studies have been conducted to investigate the role of endochitinase upon infection with oomycetes such as Phytophthora spp. From the present study it is clear that endochitinase was activated upon infection and at 72 hpi it was highly upregulated compared with the uninfected samples (0 hpi) in all rootstock varieties. Similar results were obtained in a study conducted by Mishra et al.34 who demonstrated by use of Northern blot analysis that transcripts of endochitinase were highly upregulated, reaching the highest expression 36 h after Phytophthora colocasiae elicitor treatment of taro cells.
Both glutathione-S-transferase and metallothionein are known to function as scavengers of reactive oxygen species. Antioxidant genes like glutathione-S-transferase and metallothionein displayed high basal transcript levels, particularly in the more tolerant avocado rootstocks (Dusa and R0.06). Plants rely on multiple enzymes to scavenge reactive oxygen to restore balance of toxic molecules.35 It was therefore unexpected that GTH and metallothionein were not significantly upregulated in avocado roots after P. cinnamomi treatment because scavenger molecules are known to be highly expressed during pathogenic infection, in order to remove reactive oxygen species as the presence of reactive oxygen species can be indicative of hypersensitive response activity. The highly tolerant Dusa and R0.06 displayed no significant responses for metallothionein and glutathione-S-transferase, except at 72 hpi, at which they were significantly downregulated, and for metallothionein at 24 hpi in Dusa. GTH has been shown to be upregulated in leaf tissue but downregulated in the root tissue of Coffea arabica L upon benzo(1,2,3) thiadiazole-7-carbothioic acid-s-methyl ester treatment to mimic plant disease, which supports the finding of downregulation at 72 hpi in this study. Based on our present results, neither glutathione-S-transferase nor metallothionein seem to be involved in conferring resistance in the defence response against P. cinnamomi, as the transcript levels of these genes in the highly tolerant rootstocks were not altered by pathogen infection.
Conclusion
The focus of this study was to obtain an overview of the expression patterns of selected avocado genes during P cinnamomi infection, with the intention of further understanding the mechanism of resistance observed. Because differences in gene expression are responsible for morphological and phenotypical differences, gene transcript profiles of avocado infected with P. cinnamomi, over a time period, provided evidence of genes involved in resistance and served as a basis for investigating plant-pathogen interactions and gene function.
It is highly likely that partial resistance or resistance to some pathogens, such as P. cinnamomi, is much more complex and involves the interaction of many genes at various levels and is different from those associated with R-gene mediated resistance. Identifying the genes responsible for the resistance observed in avocado rootstocks to P. cinnamomi remains a challenge. Further studies are needed to understand how resistance against P. cinnamomi is governed in avocado; this understanding will help to advance our knowledge of quantitative disease resistance in plants as well as aid in the development of markers for marker-assisted breeding for the development of resistant avocado rootstocks.
Acknowledgements
This research was funded by the Hans Merensky Foundation and the THRIP programme of the National Research Foundation of South Africa. Plant material was provided by Westfalia Technological Services and infrastructure was provided by the University of Pretoria.
Authors' contributions
J.E. contributed to the collection and analyses of the data and to the writing of the manuscript. N.v.d.B. was the project leader and assisted in the writing of the manuscript.
References
1. Zentmyer GA. Phytophthora cinnamomi and the diseases it causes. St Paul, MN: APS Press; 1980. [ Links ]
2. Pegg KG. Causes of disease. In: Broadley RH, editor. Avocado pests and disorders. Brisbane: Queensland Department of Primary Industries; 1991. p. 1-7. [ Links ]
3. Dobrowolski MP Shearer BL, Colquhoun IJ, O'Brien PA, Hardy GE. Selection for decreased sensitivity to phosphite in Phytophthora cinnamomi with prolonged use of fungicide. Plant Pathol. 2008;57(5):928-936. http://dx.doi.org/10.1111/j.1365-3059.2008.01883.x [ Links ]
4. Smith L, Dann E, Pegg K, Whiley A, Giblin F, Doogan V et al. Field assessment of avocado rootstock selections for resistance to Phytophthora root rot. Australas Plant Path. 2011;40(1):39-47. http://dx.doi.org/10.1007/s13313-010-0011-0 [ Links ]
5. Menge JA, Marais LJ. Soil environmental factors and their relationship to avocado root rot. Subtropical Fruit News. 2000;8(1-2):11-14. [ Links ]
6. Wolstenholme B. Avocado rootstocks: What do we know; are we doing enough research? South African Avocado Growers' Association Yearbook 2003. 2003;26:106-112. [ Links ]
7. Glazebrook J. Contrasting mechanisms of defence against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205-227. http://dx.doi.org/10.1146/annurev.phyto.43.040204.135923 [ Links ]
8. Kessler A, Baldwin IT. Plant responses to insect herbivory: The emerging molecular analysis. Annu Rev Plant Biol. 2002;53(1):299-328. http://dx.doi.org/10.1146/annurev.arplant.53.100301.135207 [ Links ]
9. Jackson AO, Taylor CB. Plant-microbe interactions: Life and death at the interface. Plant Cell. 1996;8(10):1651-1668. [ Links ]
10. Hardham AR. Cell biology of plant oomycete interactions. Cell Microbiol. 2007;9(1):31-39. http://dx.doi.org/10.1111/j.1462-5822.2006.00833.x [ Links ]
11. García-Pineda E, Benezer-Benezer M, Gutiérrez-Segundo A, Rangel-Sánchez G, Arreola-Cortés A, Castro-Mercado E. Regulation of defence responses in avocado roots infected with Phytophthora cinnamomi (Rands). Plant Soil. 2010;331:45-56. http://dx.doi.org/10.1007/s11104-009-0225-5 [ Links ]
12. Muñiz CA, Tovar LE, Rodriguez SV Pavia SF, Saucedo LJA, De La Cruz Espindola Barquera M, et al. Identification of avocado (Persea americana) root proteins induced by infection with the oomycete Phytophthora cinnamomi using a proteomic approach. Physiol Plant. 2012;144(1):59-72. http://dx.doi.org/10.1111/j.1399-3054.2011.01522.x [ Links ]
13. Van Den Berg N, Berger DK, Hein I, Birch PRJ, Wingfield MJ, Viljoen A. Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol Plant Pathol. 2007;8(3):333-341. http://dx.doi.org/10.1111/j.1364-3703.2007.00389.x [ Links ]
14. Kong P, Hong C, Richardson P. Rapid detection of Phytophthora cinnamomi using PCR with primers derived from the Lpv putative storage protein genes. Plant Pathol. 2003;52(6):681-693. http://dx.doi.org/10.1111/j.1365-3059.2003.00935.x [ Links ]
15. Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11(2):113-6. http://dx.doi.org/10.1007/BF02670468 [ Links ]
16. Ginzinger DG. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Experimental Hematology. 2002;30:503-12. http://dx.doi.org/10.1016/S0301-472X(02)00806-8 [ Links ]
17. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7):1-11. http://dx.doi.org/10.1186/gb-2002-3-7-research0034 [ Links ]
18. Engelbrecht J, Duong TA, Van den Berg N. Development of a nested quantitative real time PCR for detecting Phytophthora cinnamomi in Persea americana rootstocks. Plant Dis. 2013;97(8):1012-1017. http://dx.doi.org/10.1094/PDIS-11-12-1007-RE [ Links ]
19. Bowles DJ. Defence-related proteins in higher plants. Annu Rev Biochem. 1990;59(1):873-907. http://dx.doi.org/10.1146/annurev.bi.59.070190.004301 [ Links ]
20. Durrant W, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185-209. http://dx.doi.org/10.1146/annurev.phyto.42.040803.140421 [ Links ]
21. Solecka D, Kacperska A. Phenylpropanoid deficiency affects the course of plant acclimation to cold. Physiol Plant. 2003;119(2):253-262. http://dx.doi.org/10.1034/j.1399-3054.2003.00181.x [ Links ]
22. Sgarbi E, Baroni Fornasiero R, Paulino Lins A, Medeghini Bonatti P. Phenol metabolism is differentially affected by ozone in two cell lines from grape (Vitis vinifera L.) leaf. Plant Sci. 2003;165(5):951-957. http://dx.doi.org/10.1016/S0168-9452(03)00219-X [ Links ]
23. Candela ME, Alcázar MD, Espín A, Egea C, Almela L. Soluble phenolic acids in Capsicum annuum stems infected with Phytophthora capsici. Plant Pathol. 1995;44:1160123. http://dx.doi.org/10.1111/j.1365-3059.1995.tb02723.x [ Links ]
24. Koch E, Meier BM, Eiben HG, Slusarenko A. Lipoxygenase from leaves of tomato (Lycopersicon esculentum Mill.) is induced in response to plant pathogenic Pseudomonas 1. Plant Physiol. 1992;99:571-576. http://dx.doi.org/10.1104/pp.99.2.571 [ Links ]
25. Melan MA, Dong X, Endara ME, Davis KR, Ausubel FM, Peterman TK. An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 1993;101(2):441-450. http://dx.doi.org/10.1104/pp.101.2.441 [ Links ]
26. Peng YL, Shirano Y, Ohta H, Hibino T, Tanaka K, Shibata D. A novel lipoxygenase from rice. Primary structure and specific expression upon incompatible infection with rice blast fungus. J Biol Chem. 1994;269(5):3755-3761. [ Links ]
27. Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Biol. 1997;48(1):355-381. http://dx.doi.org/10.1146/annurev.arplant.48.1.355 [ Links ]
28. Weber H, Chételat A, Caldelari D, Farmer EE. Divinyl ether fatty acid synthesis in late blight diseased potato leaves. Plant Cell Online. 1999;11(3):485-494. [ Links ]
29. Rustérucci C, Montillet JL, Agnel JP Battesti C, Alonso B, Knoll A, et al. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem. 1999;274(51):36446-36455. http://dx.doi.org/10.1074/jbc.274.51.36446 [ Links ]
30. Kato T, Yamaguchi Y, Uyehara T, Yokoyama T, Namai T, Yamanaka S. Defence mechanism of the rice plant against rice blast disease. Naturwissenschaften. 1983;70(4):200-201. http://dx.doi.org/10.1007/BF01047565 [ Links ]
31. Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, Van den Elzen PJM, Cornelissen BJC. Only specific tobacco (Nicotiana tabacum) chitinases and [beta]-1,3-glucanases exhibit antifungal activity. Plant Physiol. 1993;101(3):857-863. [ Links ]
32. Erwin DC, Ribeiro OK. Phytophthora diseases worldwide. St. Paul, MN: APS Press; 1996. [ Links ]
33. Erwin DC, Bartnicki-Garcia S, Tsao PH. Phytophthora: Its biology, taxonomy, ecology, and pathology. St. Paul, MN: APS Press; 1983. [ Links ]
34. Mishra AK, Sharma K, Misra RS. Cloning and characterization of cDNA encoding an elicitor of Phytophthora colocasiae. Microbiol Res. 2010;165(2):97-107. http://dx.doi.org/10.1016/j.micres.2008.10.002 [ Links ]
35. Morita S, Kaminaka H, Masumura T, Tanaka K. Induction of rice cytosolic ascorbate peroxidase mRNA by oxidative stress; the involvement of hydrogen peroxide in oxidative stress signalling. Plant Cell Physiol. 1999;40(4):417-422. http://dx.doi.org/10.1093/oxfordjournals.pcp.a029557 [ Links ]
 Correspondence:
Correspondence:
Juanita Engelbrecht
Department of Microbiology and Plant Pathology
Forestry and Agricultural Biotechnology Institute
University of Pretoria
Private Bag X20
Hatfield 0028, South Africa
Email: Juanita.engelbrecht@fabi.up.ac.za
Received: 05 Nov. 2012
Revised: 20 Jun. 2013
Accepted: 19 Jul. 2013














