Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Science
On-line version ISSN 1996-7489
Print version ISSN 0038-2353
S. Afr. j. sci. vol.109 n.9-10 Pretoria Jan. 2013
RESEARCH LETTER
An H5N1 influenza DNA vaccine for South Africa
Elizabeth MortimerI; Inga I. HitzerothI; Amelia BuysII; Sandiswa MbewanaI; Edward P. RybickiI, III
IDepartment of Molecular and Cell Biology, University of Cape Town, Cape Town, South Africa
IINational Institute for Communicable Diseases, Johannesburg, South Africa
IIIInstitute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
ABSTRACT
The highly pathogenic avian influenza virus H5N1 is a potent pandemic threat because of its frequent transmission from birds to humans and the increasing possibility of human to human transmission. During the 2009 H1N1 pandemic it was clear that rapid influenza vaccine production is a problem worldwide. Additionally, developing countries like South Africa generally cannot produce their own influenza vaccines because the traditional egg-based vaccine production method currently employed is too lengthy and too difficult to establish. As part of an exercise aimed at exploring the feasibility of producing emergency response influenza vaccines, we investigated an experimental DNA vaccine to the H5N1 influenza virus. We focused on the virion haemagglutinin, because it elicits the primary neutralising immune response following infection. Accordingly, we developed an H5N1 DNA vaccine with full-length and truncated versions of the haemagglutinin gene, to match previously developed protein candidates. Vaccinated mice developed a strong antibody response to the haemagglutinin protein. In addition, the full-length H5 gene elicited high haemagglutination inhibition titres in mice, indicating that it has potential as a candidate pandemic vaccine for South Africa.
Keywords: DNA vaccine; highly pathogenic avian influenza virus; H5N1; haemagglutinin
Introduction
There have been five influenza pandemics over the last 125 years, with the Spanish flu in 1918 being the most notorious. The most recent ('swine flu') pandemic in 2009 was caused by influenza A H1N1, which originated in Mexico and rapidly spread globally, resulting in over 18 000 deaths.1 During this pandemic it became clear that South Africa does not have the ability to produce influenza vaccines, that we would be reliant on vaccine stocks from developed countries, and that these might be slow in coming.
The highly pathogenic avian influenza virus (HPAIV) subtype H5N1 has significant potential for causing a future pandemic, as a result of its frequent transmission from birds to mammals and continued persistence in several countries.2 There have been over 250 cases reported in humans, with deaths occurring in 60% of infected individuals. There is also the possibility of mutation and/or re-assortment of the virus to create a strain capable of enabling efficient human to human transmission; therefore, it is necessary to ensure that countries are equipped to handle a pandemic via an efficient vaccination approach.3
Current influenza vaccine production involves the use of mainly chicken egg produced trivalent inactivated vaccine. This system can take 4-6 months to produce a vaccine, and is demonstrably not capable of coping with the demand for vaccines in the event of a pandemic. Additionally, H5N1 is highly pathogenic to chickens, which has a negative effect on egg production levels.4 DNA vaccines, in contrast, can be produced in as short a time as 3 weeks.5,6 These vaccines have several advantages over the use of conventional inactivated vaccines as they can rapidly elicit both humoral and cell-mediated immune responses, and are non-infectious. DNA vaccines are heat stable and can initiate cell-mediated immune responses without the need for adjuvants.7 They are easy to construct, allowing for rapid large-scale production to meet demand in case of a pandemic.8 The potential of H5N1 DNA vaccines to protect against lethal viral challenges has also been demonstrated in animal models.9
South Africa, like other African countries, unfortunately does not have the facilities to produce conventional egg-based vaccines. When faced with the 2009 'swine flu' influenza A H1N1 pandemic, the country had to rely on the World Health Organization and developed countries for vaccine stocks.10 The 2009 H1N1 pandemic did not represent a great threat for healthy people; however, serious complications were reported for immunocompromised people. As South Africa has the highest number of human immunodeficiency virus (HIV) cases in the world,11 it is important to have an influenza pandemic contingency plan in place for when the next pandemic occurs. Ideally, developing countries like South Africa should be able to produce their own pandemic vaccine as quickly as possible. Accordingly, and as part of an effort to establish the feasibility of pandemic influenza response capability, in this paper we describe the development and testing in mice of a candidate H5 HA gene DNA vaccine in South Africa.
Materials and methods
Construction of haemagglutinin DNA vaccines
The full-length haemagglutinin (HA) gene (H5; 1704 bp) and a truncated form (H5tr; 1635 bp) lacking the 23-amino acid anchoring membrane domain (nucleotides 1597-1665 bp) of the A/Vietnam/1194/2004 strain (H5N1, GenBank accession number AY651333) were human-codon optimised and synthesised by GeneArt® (Regensburg, Germany) and called pPCR-Script_H5 and pPCR-Script_H5tr. The membrane-anchoring domain was removed from H5tr to prevent membrane insertion of the haemagglutinin. The protein produced was thus soluble, which increased production levels. The 5' ends of the HA genes were modified by means of oligonucleotide linkers by adding a Hind III restriction enzyme site and a Kozak sequence (GCCACCATGG); a BamHI site was added to the 3' end. The high-performance liquid chromatography purified oligonucleotides H5KozakF 5' CGGATCCAAGCTTACGCGTGCCAC 3' and H5KozakR 5' GTA CCACCGTGCGCATTCGAACCTAGGCCATG 3' were annealed to form a 5' end linker, while H5 3'F 5' GGCCGCATAATCTAGAGGATCCGAGCT 3' and H5 3'R 5' CGGATCCTCTAGATTAT GC 3' were annealed to form the 3' end linker. The GeneArt-provided plasmid DNAs were digested, linkers were ligated and genes were cloned into pUC 18, before sequencing with the M13F and M13R universal primers. The modified HA genes were then subcloned into the well-characterised DNA vaccine vector pTH using Hind III and Eco RI sites.12 The vector has the human cytomegalovirus enhancer and the intron A region, which resulted in high levels of vaccine protein production for a number of candidates, including HIV-1 subtype C.13
Expression of haemagglutinin DNA vaccines
Endotoxin-free DNA was prepared for both pTH_H5 and pTH_H5tr using a commercial Giga kit (Qiagen, Hilden, Germany). HA protein expression from the two constructs was verified by western blot analysis of transfected HEK 293 cells. The cells were cultured in complete medium containing Dulbecco's modified Eagle's medium (Invitrogen) with 10% foetal calf serum, 1% penicillin/streptomycin and 10 ^g/mL Fungin in T75 flasks and incubated in a 5% tissue culture 37 °C incubator until 80-90% confluency was reached. Prior to transfection, the HEK 293 cells were diluted to 0.5x106 cells and plated out in 35-mm 6-well plates. Transfection reagent (FuGENE® 6, Roche, Mannheim, Germany) was used at the recommended different reagent to DNA ratios, namely 3:1, 3:2 and 6:1. Each ratio was performed in duplicate for pTH_H5 and pTH_H5tr as well as for a negative cell control, a FuGENE® 6 only control (i.e. no DNA) and a DNA only control (i.e. no transfection reagent). After 72 h, the HEK cells were harvested with cold Dulbecco's phosphate buffered saline (PBS, Sigma) and were lysed with 1 x chloramphenicol acetyltransferase lysis buffer (Roche).
Lysed cell samples and supernatant were assayed for protein expression by loading the sample on a 10% SDS-polyacrylamide (PAGE) gel. The transfer of proteins from gels to nitrocellulose membranes was done by means of semi-dry electroblotting (Bio-Rad). Rabbit anti-H5N1 polyclonal antibody (US Biological, GenBank accession number AAT6166) was used at a 1:1000 dilution together with a secondary alkaline phosphatase-labelled goat anti-rabbit antibody (Sigma) at a 1:7000 dilution. NBT/BCIP solution (Roche) was used for detection. We determined that the optimum transfectant to DNA ratios for H5 and H5tr were 6:1 and 3:1, respectively. This transfection experiment was repeated twice to confirm results.
Mouse immunisation
A total of 30 healthy 7-week-old female Balb/c mice were equally divided into three groups. Each group received one of the following: (1) pTH_H5 DNA vaccine, (2) pTH_H5tr DNA vaccine or (3) PBS (a negative control group). A total dose of 100 pTH_H5 and pTH_H5tr endotoxin-free DNA in 100 was administered intramuscularly to mice - 50 into each anterior tibialis muscle - after mice received an intraperitoneal anaesthetic mixture of ketamine (120 mg/kg) and xylazine (16 mg/kg, Rompun®). Four doses were administered at 2-week intervals. The mice were pre-bled (100 ^L) 3 days before vaccination was initiated. Final blood samples were taken 2 weeks after the fourth vaccine dose. The animal experiments were approved by the University of Cape Town's Animal Ethics Committee (HSFAEC 009001). At the end of the experiment, sera were collected and stored at -20 °C for further processing.
Serological assays
Western blots were performed on in-house plant-produced H5 and H5tr protein samples (A/Vietnam/1194/2004)14 separated on 10% SDS-PAGE gels. Following semi-dry blotting, membranes were cut into individual strips and incubated in 1:200 dilutions of sera from immunised mice. A positive control was also included that used the primary antibody (rabbit anti-H5N1 polyclonal antibody, US Biologicals, Swampscott, MA, USA) at a 1:500 dilution followed by the secondary goat anti-rabbit antibody (Sigma-Aldrich, Johannesburg, South Africa).
Haemagglutination inhibition (HI) tests were performed at the National Institute for Communicable Diseases (NICD, Johannesburg, South Africa). The mice sera were treated with a receptor destroying enzyme (RDE II, Denka Seiken) to a dilution of 1:10 (1 part serum + 3 parts RDE, incubated overnight at 37 °C, heat inactivated at 56 °C for 30 min and another 6 volumes of saline added). Following this, serial dilutions of the sera were mixed with a standard 4HAU/25-^L dose of Influenza A H5N1 antigen received from the Centers for Disease Control and Prevention (CDC, Atlanta, GA, USA). Following incubation, a 0.5% suspension of turkey red blood cells was used as an indicator system to assess inhibition of haemagglutination. Influenza A (H3N2) and Influenza A (H1N1) antigens (CDC) were used as negative controls to check for cross-reaction. To test for significant differences in antibody titres between the two treatment groups (pTH_H5 and pTH_H5tr), we performed an independent t-test, after confirming the assumption of normality with Shapiro-Wilk tests for each group (p>0.05).
Results and discussion
H1N1 and H5N1 influenza viruses are of keen current interest - H1N1 because of the role it played in the recent influenza pandemic and H5N1 because of its potential to cause another pandemic.15,16 DNA vaccines continue to be an attractive option for vaccine production in the event of a pandemic influenza outbreak as a result of their potential for rapid, large-scale development. Influenza DNA vaccines have been shown to be as effective as live vaccines, which can protect mice against viral challenges, and have entered advanced human trials.6,8,17
We explored the possibility of making a HA DNA vaccine in response to a future pandemic in southern Africa, using an established DNA vaccine vector which has been used in primate and human studies in the UK and in human trials in South Africa,13,18,19 and which contained the full-length and truncated forms of a H5N1 HA gene. Both HA variants were expressed in HEK 293 cell cultures (results not shown). After confirmation of expression, mice were subsequently vaccinated with four doses of these potential DNA vaccines. Two weeks following the last vaccination, the sera were collected. Western blots were used to determine if H5-specific antibodies were elicited in the mice sera following immunisation. Sera against both the full-length and truncated vaccine candidates bound strongly to in-house plant-produced H5 (Figure 1) from the same strain (A/Vietnam/1194/2004) as the HA DNA vaccines. As expected, the sera of mice inoculated with PBS (negative control) contained no H5-specific antibodies. The positive control (Lane 19) appears weaker than the rest as the commercial antibody targets 14 amino acids in the middle of the HA protein of a strain (AAT76166) different from the one used in this study. H5-specific antibodies from the sera of mice vaccinated with pTH_H5 and pTH_H5tr appear to be mostly strain specific, even though cross-reaction weakly occurs, as indicated by the positive control. Ideally, cross-reaction with other influenza strains will make the candidate vaccines more suitable for broader influenza strain protection. However, as a pandemic usually starts off with a single viral strain, these results are in line with producing a candidate pandemic vaccine.
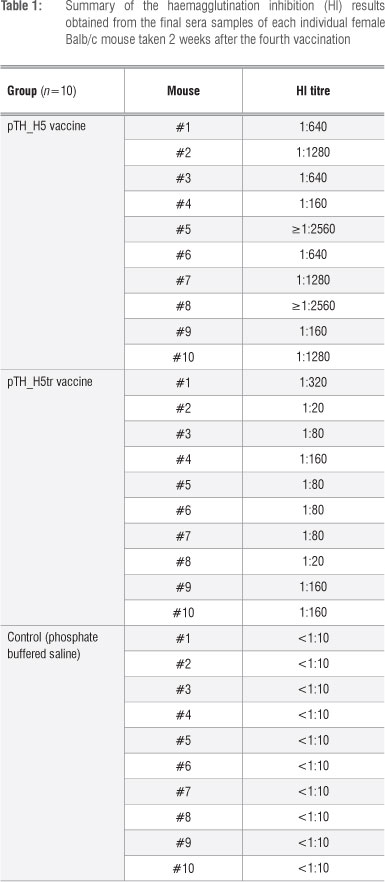
The sera were further tested for the ability to inhibit haemagglutination. The HI results (Table 1) indicated that sera from all 10 mice vaccinated with the full-length HA gene (pTH_H5) had HI titres ranging from 1:160 to >1:2560. These HI titres were therefore considerably greater than 1:40, which is deemed to be the benchmark for protection against influenza in humans.20,21 In contrast, the truncated vaccine (pTH_H5tr) produced lower titres that ranged from 1:20 to 1:320. Thus, 8 of 10 mice had HI titres greater than 1:40. The lower HI values could be because the truncated HA form cannot be membrane-bound, which probably reduces its immunogenicity. Antibody titres were also significantly higher (f=3.645; df=18; p=0.002) for pTH_H5 (1120.00±866.56, mean±sd) than for pTH_H5tr (116.00±88.84). Thus, the full-length HA (H5) DNA candidate vaccine proved to be more immunogenic than its truncated counterpart. These findings are in line with our previous results when comparing our plant-produced H5 and H5tr subunit candidate vaccines.14
It has been shown that HA DNA vaccines can protect when there is a high concentration of HI and neutralising antibodies present.22 The potential of H5N1 DNA vaccines has been highlighted by studies that indicated a reduction of morbidity and mortality of test animals during a lethal H5N1 viral challenge; however, DNA vaccines generally elicit only weak immune responses in humans.9 An alternative strategy to prevent pandemic outbreaks that has been proposed is the use of a DNA prime followed by an inactivated vaccine boost, as this regime elicits good immune responses, and one could vaccinate people with the DNA vaccine before an outbreak as part of seasonal influenza vaccination.23 To this end, we have also developed a plant-made H5 HA subunit vaccine candidate, which elicits good serum responses in mice and chickens.14
The development of a universal influenza vaccine is ideal and has been investigated in an attempt to prevent pandemics. Generally the focus is on the highly conserved ectodomain of the matrix-2 protein (M2e) of influenza subtype A viruses or haemagglutinin's conserved stalk domain (for a review see Pica and Palese24). For example, a DNA vaccine study has shown that the fusion of M2e to H1N1 HA DNA or the mixture of M2e and HA DNA improved cross-protection against H5N2 in mice, in contrast to immunisations with HA and M2e DNA on their own.25 Also, to generate a broadly protective DNA swine vaccine, M2e was fused to H3 HA (consensus sequence), as well as to a CTL epitope to H3 HA. The DNA vaccines generated offered complete homologous strain protection and some heterologous protection in mice.23 It is therefore worth considering, in terms of future work, determining if there is an increase in immunogenicity and cross-protection with the addition of M2e to our H5 and H5tr HA constructs. We have already investigated the plant production of M2e in fusion with another protein; the result could also serve as a combination vaccine (E. Mortimer, I.I. Hitzeroth, S. Mbewana and E.P Rybicki, unpublished results).
DNA vaccines can be stockpiled and can be manufactured in a matter of weeks: if other HA genes need to be cloned, all the methodologies are established, and the vaccine could be ready within weeks of notification. By implementing a DNA vaccine platform, South Africa can respond rapidly and potentially remain self-sufficient during a potential influenza pandemic.
Conclusion
The H5 DNA vaccine we prepared shows excellent potential in mice; it produced good humoral responses and elicited a good HI response. We believe this work has further helped us to establish a basis for 'rapid response' influenza vaccine production in South Africa in order to develop pandemic preparedness.
Acknowledgements
This research was funded by the Poliomyelitis Research Foundation. The National Research Foundation (South Africa) provided E.M. with a postdoctoral bursary. We thank Fiona Tanzer for designing the oligonucleotide linkers; Desiree Bowers for providing the HEK 293 cell line; Rodney Lucas and Enid Shephard for aiding with animal experiments; Thomas Hanke for supplying us with the mammalian expression vector pTH; Aleyo Chabeda for assisting with the manuscript; Maya Pfaff for assisting with statistical analyses; and two anonymous reviewers for their valuable input.
Authors' contributions
E.M. participated in the design of the study, carried out expression and animal experiments, performed the serum analysis and drafted the manuscript; I.I.H. designed and coordinated the study, helped draft the manuscript and revised the paper; A.B. performed the haemagglutination inhibition assays; S.M. participated in the mouse experiments and serum analysis; and E.P.R. conceived the study, made the H5N1 strain selection, participated in the design of the study and in the drafting of the manuscript. All authors read and approved the final manuscript.
References
1. Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr., Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357-360. http://dx.doi.org/10.1126/science.1186430 [ Links ]
2. Kieny MP Costa A, Hombach J, Carrasco P Pervikov Y Salisbury D, et al. A global pandemic influenza vaccine action plan. Vaccine. 2006;24:6367-6370. http://dx.doi.org/10.1016/j.vaccine.2006.07.021 [ Links ]
3. Webster RG, Govorkova EA. H5N1 influenza-continuing evolution and spread. N Engl J Med. 2006;355:2174-2177. http://dx.doi.org/10.1056/NEJMp068205 [ Links ]
4. Hu AY Weng TC, Tseng YF, Chen YS, Wu CH, Hsiao S, et al. Microcarrier-based MDCK cell culture system for the production of influenza H5N1 vaccines. Vaccine. 2008;26:5736-5740. http://dx.doi.org/10.1016/j.vaccine.2008.08.015 [ Links ]
5. Forde GM. Rapid-response vaccines - Does DNA offer a solution? Nat Biotechnol. 2005;23:1059-1062. http://dx.doi.org/10.1038/nbt0905-1059 [ Links ]
6. Pandey A, Singh N, Mittal SK. Egg-independent vaccine strategies for highly pathogenic H5N1 influenza viruses. Hum Vaccin. 2010;6:178-188. http://dx.doi.org/10.4161/hv.6.2.9899 [ Links ]
7. Sharma AK, Khuller GK. DNA vaccines: Future strategies and relevance to intracellular pathogens. Immunol Cell Biol. 2001;79:537-546. http://dx.doi.org/10.1046/j.1440-1711.2001.01044.x
8. Yager EJ, Dean HJ, Fuller DH. Prospects for developing an effective particle-mediated DNA vaccine against influenza. Expert Rev Vaccines. 2009;8:1205-1220. http://dx.doi.org/10.1586/erv.09.82
9. Chen MW, Cheng TJ, Huang Y Jan JT, Ma SH, Yu AL, et al. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc Natl Acad Sci USA. 2008;105:13538-13543. http://dx.doi.org/10.1073/pnas.0806901105 [ Links ]
10. World Health Organization (WHO). Report of the WHO Pandemic Influenza A(H1N1) Vaccine Deployment Initiative. Geneva: WHO; 2011. [ Links ]
11. UNAIDS. World AIDS Day report. Geneva: UNAIDS; 2011. [ Links ]
12. Hanke T, Blanchard TJ, Schneider J, Hannan CM, Becker M, Gilbert SC, et al. Enhancement of MHC class I-restricted peptide-specific T-cell induction by a DNA prime/MVA boost vaccination regime. Vaccine. 1998;16:439-445. http://dx.doi.org/10.1016/S0264-410X(97)00226-0 [ Links ]
13. Burgers WA, Chege GK, Muller TL, Van Harmelen JH, Khoury G, Shephard EG, et al. Broad, high-magnitude and multifunctional CD4+ and CD8+ T-cell responses elicited by a dNa and modified vaccinia Ankara vaccine containing human immunodeficiency virus type 1 subtype C genes in baboons. J Gen Virol. 2009;90:468-180. http://dx.doi.org/10.1099/vir.0.004614-0
14. Mortimer E, Maclean JM, Mbewana S, Buys A, Williamson AL, Hitzeroth II, et al. Setting up a platform for plant-based influenza virus vaccine production in South Africa. BMC Biotechnology. 2012;12:14. http://dx.doi.org/10.1186/1472-6750-12-14
15. Murray CJL, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918-20 pandemic: A quantitative analysis. Lancet. 2006;368:2211-2218. http://dx.doi.org/10.1016/S0140-6736(06)69895-4
16. Fraser C, Donnelly CA, Cauchemez S, Hanage WP Van Kerkhove MD, Hollingsworth TD, et al. Pandemic potential of a strain of Influenza A (H1N1): Early findings. Science. 2009;324:1557-1561. http://dx.doi.org/10.1126/science.1176062
17. Patel A, Gray M, Li Y Kobasa D, Yao X, Kobinger GP. Co-administration of certain DNA vaccine combinations expressing different H5N1 influenza virus antigens can be beneficial or detrimental to immune protection. Vaccine. 2012;30:626-636. http://dx.doi.org/10.1016/j.vaccine.2011.11.017
18. Hanke T, Goonetilleke N, McMichael AJ, Dorrell L. Clinical experience with plasmid DNA- and modified vaccinia virus Ankara-vectored human immunodeficiency virus type 1 clade A vaccine focusing on T-cell induction. J Gen Virol. 2007;88:1-12. http://dx.doi.org/10.1099/vir.0.82493-0
19. Knudsen ML, Mbewe-Mvula A, Rosario M, Johansson DX, Kakoulidou M, Bridgeman A, et al. Superior induction of T cell responses to conserved HIV-1 regions by electroporated alphavirus replicon DNA compared to that with conventional plasmid DNA vaccine. J Virol. 2012;86:4082-4090. http://dx.doi.org/10.1128/JVI.06535-11
20. Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond). 1972;70:767-777. http://dx.doi.org/10.1017/S0022172400022610
21. Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis. 2011;204:1879-1885. http://dx.doi.org/10.1093/infdis/jir661
22. Ulmer JB. Influenza DNA vaccines. Vaccine. 2002;20(suppl 2):S74-S76. http://dx.doi.org/10.1016/S0264-410X(02)00136-6 [ Links ]
23. Wang S, Hackett A, Jia N, Zhang C, Zhang L, Parker C, et al. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PLoS One. 2011;6:e28757. http://dx.doi.org/10.1371/journal.pone.0028757
24. Pica N, Palese P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu Rev Med. 2013;64:189-202. http://dx.doi.org/10.1146/annurev-med-120611-145115
25. Park KS, Seo YB, Lee JY Im SJ, Seo SH, Song MS, et al. Complete protection against a H5N2 avian influenza virus by a DNA vaccine expressing a fusion protein of H1N1 HA and M2e. Vaccine. 2011;29:5481-5487. http://dx.doi.org/10.1016/j.vaccine.2011.05.062
 Correspondence:
Correspondence:
Inga Hitzeroth
Department of Molecular and Cell Biology
University of Cape Town
Rondebosch 7701, South Africa
Email: inga.hitzeroth@uct.ac.za
Received: 31 Oct. 2012
Revised: 11 Apr. 2013
Accepted: 12 Apr. 2013














