Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Science
versión On-line ISSN 1996-7489
versión impresa ISSN 0038-2353
S. Afr. j. sci. vol.104 no.7-8 Pretoria sep./ago. 2008
RESEARCH LETTERS
A broad host range reporter plasmid for the analysis of divergent promoter regions
Meesbah Jiwaji*, ª ; Gwynneth Felicity Matcherb; Rosemary Ann Dorrington
Department of Biochemistry, Microbiology and Biotechnology, Rhodes University, P.O. Box 94, Grahamstown 6140, South Africa
ABSTRACT
Although many vectors exist for Escherichia coli and closely related species, there are few broad host range vectors that can be conjugated into a large variety of Gram-negative bacteria. We have constructed a broad host range vector, pMJ445, that facilitates the analysis of divergent promoters in Gram-negative bacteria. The vector was validated using two intergenic regions derived from gene clusters involved in hydantoin hydrolysis, from the environmental isolates Pseudomonas putida and Agrobacterium tumefaciens. The DNA sequences analysed were capable of activating expression of the reporter enzymes, β-glucuronidase and β-galactosidase, present on pMJ445, indicating the presence of divergent promoters in the sequences selected. In addition, we demonstrated that pMJ445 can be applied to gene regulation studies.
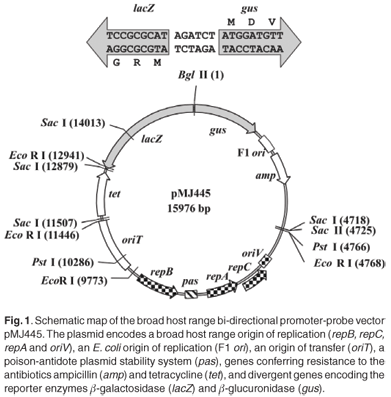
Two major difficulties with studying gene regulation in environmental microbial isolates are the lack of stable, broad host range promoter-probe vectors and inefficient methods for introducing recombinant plasmids into these strains. There are even fewer vectors for studying transcriptional activity directed by divergent or bi-directional promoter systems. We have constructed a promoter-probe plasmid, pMJ445 (GenBank Accession No. EU250578), which enables the simultaneous assay of transcriptional activity directed by divergent promoter sequences via β-galactosidase and β-glucuronidase reporter activity in a wide variety of Gram-negative bacteria. pMJ445 is based on the broad host range IncQ replicon derived from pTF-FC2.1 The pTF-FC2 replicon is small (4.9 kb) and its host range includes Escherichia coli, Pseudomonas aeruginosa, Acidithiobacillus ferrooxidans and Agrobacterium tumefaciens.1,2 Furthermore, the pTF-FC2 replicon maintains the plasmid at a low copy number (12–15 copies per cell)3 and the presence of a poison-antidote system ensures stability of the plasmid in the host cells.4 To facilitate easy propagation and extraction of plasmid DNA in high concentrations in E. coli DH5α, pMJ445 carries the F1 ori as well as genes conferring resistance to ampicillin (E. coli DH5α) and tetracycline (broad host range). pMJ445 also carries the RK2 oriT gene, which allows for efficient mobilization into strains of interest by conjugation in addition to chemical transformation and electroporation. Promoter sequences, including native ribosome binding sites, can be readily inserted into the single Bgl II site, located between the divergently orientated lacZ and gus ORFs (Fig. 1), for analysis. Microbial hydantoin-hydrolysing enzyme systems have important industrial applications in the biocatalytic production of optically pure D- and L-amino acids, which are used in the synthesis of antibiotics, anti-inflammatory and anti-viral drugs.5 Hydantoin hydrolysis occurs via two steps. First, the 5-monosubstituted-hydantoin is cleaved by hydantoinase or dihydropyrimidinase to produce the N-carbamylamino acid, which is in turn converted to the corresponding amino acid by an N-carbamoylase or β-ureidopropionase. In Pseudomonas putida, a dihydropyrimidinase and β-ureidopropionase (encoded by dhp and bup genes, respectively) are responsible for hydantoin hydrolysis.6 In Agrobacterium species, hydantoin hydrolysis is catalysed by a hydantoinase and N-carbamoylase (encoded by hyuH and hyuC genes, respectively).7,8 In both cases, the hyuH-hyuC and dhp-bup genes are arranged divergently,6–8 with an additional gene, encoding a putative permease, located upstream of and in the same orientation as bup in P. putida strain RU-KM3s.6 Hydantoin-hydrolysing activity is tightly controlled by a complex regulatory network. The enzymes are expressed in early stationary growth phase and activity is induced when cells are grown in the presence of hydantoin.9–11 In Agrobacterium tumefaciens, hydantoin hydrolysis is also subject to nitrogen catabolite repression,12 whereas hydantoin hydrolysis in P. putida RU-KM3s is regulated by carbon catabolite repression (CCR).6 The molecular basis for these regulatory pathways is unknown. This provides an opportunity to test the broad host range plasmid pMJ445 in two diverse genera of Gram-negative bacteria and to elucidate the mechanisms involved in the regulation of hydantoin hydrolysis in A. tumefaciens RU-AE01 and P. putida RU-KM3s.
Materials and methods
Construction of the broad host range vector involved six steps. 1) The RP4 oriT–tet–MB1 oriV fragment was excised from pTnmodOTc13 with the restriction enzymes Pvu II and Sal I and inserted between the Sma I and Sal I sites of pT7-7.14 2) The RP4 oriT–tet–MB1 oriV fragment was then excised from this intermediate construct with Csp 451 and Kpn I and inserted between the Cla I and Kpn I sites of pTV100,15 generating pJAS13. 3) Plasmid pJAS13 contained a Bgl II restriction site that was important in the future cloning strategy. This restriction site was deleted by digesting pJAS13 with Bgl II, filling in the termini with Klenow enzyme and re-circularizing the plasmid (pMJ417). 4) The gus–RU-AE01 promoter–lacZ fragment was amplified from pMJ25819 using the Expand High Fidelity PCR system (Roche) and the primers MJ65 (ggt acc TCA TTG TTT GCC TCC CTG CTG CG) and MJ66 (ccg cgg GGA TTA TTT TTG ACA CCA GAC CAA CTG GTA ATG GTA G), which introduced Kpn I and Sac II sites, respectively (shown in lower case in the primer sequence), and inserted into pGEM-T-Easy (Promega), generating pMJ383. 5) The gus–RU-AE01 promoter–lacZ fragment was excised from pMJ383 with the restriction enzymes Kpn I and Sac II and the termini filled in with Klenow enzyme. Similarly, pMJ417 was digested with Cla I and Kpn I and the termini filled in with Klenow enzyme. These blunt-end DNA fragments were ligated and pMJ441, which encoded gus–RU-AE01 promoter–lacZ, RP4 oriT–tet and the broad origin of replication (repB, repA, repC and oriV), was isolated. 6) Finally, pMJ441 was digested with Bgl II, to excise the RU-AE01 promoter fragment, and the DNA backbone re-circularized to produce the broad host range promoter plasmid pMJ445. This broad host range promoter-probe vector was validated using restriction mapping, PCR analysis with primers designed to amplify across the regions where the different fragments were annealed, followed by DNA sequencing.
The divergent promoters and native ribosome binding sites of the hyuH-hyuC and dhp-bup gene clusters from A. tumefaciens RU-AE01 and P. putida RU-KM3s were used to validate the application of pMJ445 as a broad host range promoter-probe vector. The region between the A. tumefaciens hyuH-hyuC coding sequences was amplified using the Expand High Fidelity PCR system and primers MJ9 (aga tct AAA GCA GCT CTC AGG GTT GAT G) and MJ10 (aga tct GAA CCT TTG CTC CTT CGA TAG TTA AT), which introduced flanking Bgl II sites (shown in lower case in the primer sequence), and the PCR product inserted between the reporter genes in pMJ445. Likewise, the P. putida dhp-bup promoter region was amplified using primers GFM38 (aga tct GGG GCC TTC TCC AGA TTT TT) and GFM39 (aga tct GCC GTC TTC CTC GCA G) and the PCR product inserted into the Bgl II site of pMJ445. pMJ441 carries the A. tumefaciens hyuH-promoter upstream of gus and the hyuC-promoter upstream of lacZ, whereas pMJ449 has the P. putida dhp-promoter upstream of gus and the bup-promoter upstream of lacZ.
A. tumefaciens RU-AE01 cells were transformed by electroporation16 with a transformation efficiency of 104 colonies per µg plasmid DNA. Plasmids were introduced into P. putida RU-KM3s cells by tri-parental mating during which plasmids were mobilized from E. coli DH5α cells with the aid of E. coli HB101 cells containing the helper plasmid pRK2013.6 Mating frequencies of 10–5–10–4 transconjugants per donor were routinely observed. For enzyme assays, cultures were grown at 28°C in 100 ml Nutrient Broth (NB) (Biolab) with or without inducer (1% hydantoin, Sigma) and 1% succinate (United Scientific) (to detect CCR). A. tumefaciens and P. putida cells were harvested in late stationary (OD600 nm = 4.5–5.0) and early stationary growth phase (OD600 nm = 2.5–3.0), respectively. Cells were collected by centrifugation (5000 rpm for 10 min at 10°C in a Beckman centrifuge in a JA14 rotor), washed in 0.1 M phosphate buffer [pH 8.0 (P. putida) or pH 9.0 (A. tumefaciens)] and resuspended in 0.1 M phosphate buffer [pH 8.0 (P. putida) or pH 9.0 (A. tumefaciens)] at 20 mg/ml wet cell mass. The cells were subsequently incubated in the presence of the substrate hydantoin (50 mM) or N-carbamylglycine (25 mM) (Sigma) at 40°C, 100 rpm (to ensure constant mixing of the reagents in the assay), for 6 h (A. tumefaciens) or 3 h (P. putida). The cells in the resting cell reactions were pelleted by centrifugation (13 000 rpm at room temperature in a Heraeus microfuge) and the supernatant analysed for N-carbamylamino acids or amino acids by Ehrlich's or Ninhydrin colorimetric assays, respectively.17,18 Hydantoinase and N-carbamoylase enzyme assays in A. tumefaciens cells, dihydropyrimidinase and β-ureidopropionase enzyme assays in P. putida cells, and β-glucuronidase assays in both genera were conducted as described previously.6,9,19 P. putida cells were disrupted by sonication prior to β-galactosidase assays.20 Hydantoinase activity is reported as the total (in µmol/ml) N-carbamoylglycine and glycine produced from hydantoin as a substrate by resting cells and N-carbamoylase activity is reported as the amount of glycine (in µmol/ml) produced from N-carbamoylglycine as a substrate. All biocatalytic assays were independently repeated at least three times with freshly cultured cells.
Results
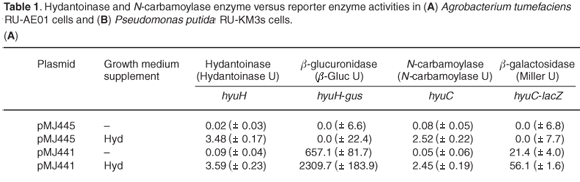
Hydantoinase and N-carbamoylase activities were undetectable in A. tumefaciens cells grown in NB but there was a 3.5-fold and 2.5-fold increase in hydantoinase and N-carbamoylase activity, respectively, when A. tumefaciens cells were grown in the presence of hydantoin (Table 1A). No β-glucuronidase or β-galactosidase activity was detected in A. tumefaciens cells containing the vector pMJ445 (no promoter), indicating a lack of endogenous reporter enzyme activity under analysis conditions. A 3.5-fold increase in hyuH-directed β-glucuronidase activity and a 2.5-fold increase in the hyuC-promoter derived β-galactosidase activity were observed in A. tumefaciens (pMJ441) cells grown in NB containing hydantoin. The correlation between the hydantoinase and N-carbamoylase enzyme activities with those of the reporter enzymes, β-glucuronidase and β-galactosidase, indicated that induction of hydantoin hydrolysis in A. tumefaciens cells occurs at the transcriptional level.
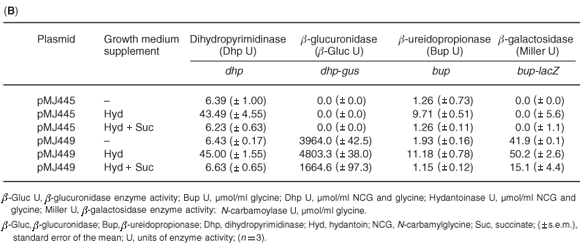
In P. putida (pMJ445), dihydropyrimidinase activity was induced 7-fold by growth in the presence of hydantoin (Table 1B), while the addition of succinate resulted in a 6.8-fold decrease in dihydropyrimidinase activity. A small increase was observed in dhp-directed β-glucuronidase activity in P. putida (pMJ449) cells grown in hydantoin, while the addition of succinate resulted in a 2.9-fold decrease of β-glucuronidase activity. As with the dihydropyrimidinase, β-ureidopropionase activity was induced 5.8-fold when P. putida (pMJ445) cells were grown in NB containing hydantoin and the presence of succinate resulted in a 9.7-fold reduction in activity. A small but important increase in bup-directed β-galactosidase activity was detected in P. putida (pMJ449) cells grown in hydantoin with a 3.3-fold decrease in activity in cells grown in both hydantoin and succinate. The results suggested that the changes in dihydropyrimidinase and β-ureidopropionase activities observed in P. putida cells grown in the presence of hydantoin or hydantoin and succinate can be attributed, at least in part, to the regulation of transcriptional activation by the dhp-bup promoters. The level of induction or repression of the reporter enzymes, β-glucuronidase and β-galactosidase, was not as high as that observed for the native enzymes in P. putida. This may be due to the effect of plasmid copy number (12–15 copies per cell versus one chromosomal copy each of dhp and bup). Alternative explanations may be differences in the half-lives of the reporter enzymes versus the hydantoin-hydrolysing enzymes or the possibility of post-translational modification of the dihydropyrimidinase and β-ureidopropionase enzymes in P. putida.
These results demonstrate that pMJ445 can be used to analyse divergent regulatory regions simultaneously. We were able to introduce DNA fragments up to 1 kb between the genes for the reporter enzymes. We also successfully applied the plasmid pMJ445 in deletion analyses and site-directed mutagenesis to identify the regulatory elements in the dhp-bup and the hyuH-hyuC promoter regions (data not shown). The ability to mobilize pMJ445 and its derivatives efficiently into environmental isolates demonstrates the flexibility of this broad host range plasmid. Finally, the stability of pMJ445 and the ease of use make this vector a valuable tool in the study of regulatory regions in a wide variety of Gram-negative bacterial strains.
This research was funded by a grant from the National Research Foundation (GUN 2073134) and the DACST Innovation Fund (Grant No. 41241). We thank Doug Rawlings (University of Stellenbosch) for providing the pTF-FC2 replicon. M.J. was supported by a scholarship from the Andrew Mellon Foundation.
1. Rawlings D.E. and Tietze E. (2001). Comparative biology of the IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65, 481–496. [ Links ]
2. Dorrington R.A. and Rawlings D.E. (1990). Characterization of the minimum replicon of the broad host range plasmid pTF-FC2 and similarity between pTF-FC2 and the IncQ plasmids. J. Bacteriol. 172, 5697–5705. [ Links ]
3. Dorrington R.A. and Rawlings D.E. (1989). Identification and sequence of the basic replication region of a broad host range plasmid isolated from Thiobacillus ferrooxidans. J. Bacteriol. 171, 2735–2739. [ Links ]
4. Smith A.S.G. and Rawlings D.E. (1997). The poison-antidote stability system of the broad host range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol. Microbiol. 26, 961–970. [ Links ]
5. Burton S.G. and Dorrington R.A. (2004). Hydantoin-hydrolyzing enzymes for the enantioselective production of amino acids: new insights and applications. Tetrahedron Asymmetry 15, 2737–2741. [ Links ]
6. Matcher G.F., Burton S.G. and Dorrington R.A. (2004). Mutational analysis of the hydantoin hydrolysis pathway in Pseudomonas aeruginosa RU-KM3s. Appl. Microbiol. Biotechnol. 65, 391–400. [ Links ]
7. Grifantini R., Galli G., Carpani G., Pratesi C., Frascotti G. and Grandi C. (1998). Efficient conversion of 5-substituted hydantoins to D-α-amino acids using recombinant Escherichia coli strains. Microbiology 144, 947–954. [ Links ]
8. Hils M., Munch P., Altenbuchner J., Syldatk C. and Mattes R. (2001). Cloning and characterization of genes from Agrobacterium sp. IP I-671 involved in hydantoin degradation. Appl. Microbiol. Biotechnol. 57, 680–688. [ Links ]
9. Hartley C.J., Kirchmann S., Burton S.G. and Dorrington R.A. (1998). Production of D-amino acids from D, L-substituted hydantoins by an Agrobacterium tumefaciens strain and isolation of a mutant with inducer-independent expression of hydantoin-hydrolyzing activity. Biotechnol. Lett. 20, 707–711. [ Links ]
10. Meyer P. and Runser S. (1993). Efficient production of the industrial biocatalysts hydantoinase and N-carbamyl amino acid amidohydrolase: Novel non-metabolisable inducers. FEMS Microbiol. Lett. 109, 67–74. [ Links ]
11. Runser S., Chinski N. and Ohleyer E. (1990). D-p-hydroxyphenylglycine production from D, L-5-p-hydroxyphenylhydantoin by Agrobacterium sp. Appl. Microbiol. Biotechnol. 33, 382–388. [ Links ]
12. Hartley C.J., Manford F.P., Burton S.G. and Dorrington R.A. (2001). Over-production of hydantoinase and N-carbamoylamino acid amidohydrolase enzymes by regulatory mutants of Agrobacterium tumefaciens. Appl. Microbiol. Biotechnol. 57, 43–49. [ Links ]
13. Dennis J. and Zylstra G. (1998). Plasposons: modular self cloning mini-transposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64, 2710–2715. [ Links ]
14. Tabor S. and Richardson C. (1987). DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl Acad. Sci. USA 84, 4767–4771. [ Links ]
15. Dorrington R., Bardien S. and Rawlings D. (1991). The broad host range plasmid pTF-FC2 requires a primase-like protein for autonomous replication in Escherichia coli. Gene 108, 7–14. [ Links ]
16. Shaw C.H. (1995). Introduction of cloning plasmids into Agrobacterium tumefaciens. Methods Mol. Biol. 49, 33–37. [ Links ]
17. Morin A., Hummel W., Schutte H. and Kula M-R. (1986). Characterization of hydantoinase from Pseudomonas fluorescens strain DSM84. Biotechnol. Appl. Biochem. 8, 564–574. [ Links ]
18. Plummer D. (1987). In Practical Biochemistry, 3rd edn, p. 158. Wiley-Interscience, New York. [ Links ]
19. Jiwaji M. (2007). Regulation of hyu gene expression in Agrobacterium tumefaciens strains RU-AE01 and RU-OR, pp. 52–52 and 77–78. Ph.D. thesis, Rhodes University, Grahamstown. [ Links ]
20. Miller J. (1992). In A Short Course in Bacterial Genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, New York. [ Links ]
Received 6 June. Accepted 18 August 2008.
* Author for correspondence. E-mail: m.jiwaji@bio.gla.ac.uk
a Present address: Department of Biochemistry and Molecular Biology, University of Glasgow, Glasgow G12 8QQ, Scotland, U.K
b Present address: Department of Microbiology, Stellenbosch University, Private Bag X1, Matieland 7602, South Africa