Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Science
versão On-line ISSN 1996-7489
versão impressa ISSN 0038-2353
S. Afr. j. sci. vol.103 no.7-8 Pretoria Jul./Ago. 2007
RESEARCH IN ACTION
Hyper-resistance to arsenic in bacteria isolated from an antimony mine in South Africa
E. Botes; E. Van Heerden‡; D. Litthauer
Department of Biotechnology, University of the Free State, P.O. Box 339, Bloemfontein 9300, South Africa
ABSTRACT
Soil and water sites were sampled at a South African antimony mine with elevated levels of arsenic due to the refining process. Enriched media yielded two pure bacterial cultures able to grow in both arsenite and arsenate. These were identified as Stenotrophomonas maltophilia SA Ant 15 and Serratia marcescens SA Ant 16. Stenotrophomonas maltophilia SA Ant 15 was resistant to 10 mmol l–1 arsenite and 20 mmol l–1 arsenate, whereas S. marcescens SA Ant 16 grew in 15 mmol l–1 arsenite and in up to 500 mmol l–1 arsenate, making it the most arsenic-resistant organism described to date. During growth, addition of arsenate or arsenite anions adversely affected biomass production and maximum specific growth rate and, in some instances, longer lag phases were induced. Reduction of arsenate to arsenite partly accounted for the high tolerance of the bacteria to arsenate. Our results suggest the use of these hyper-resistant bacteria as remediation agents in areas where arsenic contamination is prohibitively high.
Introduction
Arsenic is widely distributed throughout the earth's crust, ranging from trace levels up to hundreds of milligrams per kilogram. Sources of contamination include erosion of local rocks, industrial effluents, various commercial processes and combustion of fossil fuels.1 Arsenic can exist in several oxidation states, but under aerobic conditions arsenate [As(V)] predominates, while in reducing environments arsenite [As(III)] is the dominant species. A change in the oxidation state of arsenic alters the solubility of the oxyanion, with potential applications in the bio-remediation industry.2 Although As(III) is considered to be up to 1000 times more toxic,3 and is generally more mobile, than As(V),2 it readily forms precipitates with metal sulphides4 or hydrous oxides of iron5 and can therefore be removed from solution. Microorganisms have a variety of ways to cope with high levels of arsenic, ranging from reduced uptake,6 methylation7 and adsorption,8 to dissimilatory arsenate respiration.9 The most widespread and best-described mechanism is reduction of arsenate and extrusion of the resulting arsenite by soluble reductases and membrane-associated pumps.10–13 The aim of the study reported here was to isolate and identify bacteria from an arsenic-impacted environment, to determine the level of arsenic resistance and to establish if resistance is due to the reduction of arsenate to arsenite.
Materials and methods
See Appendix
Results
Enrichments and identification
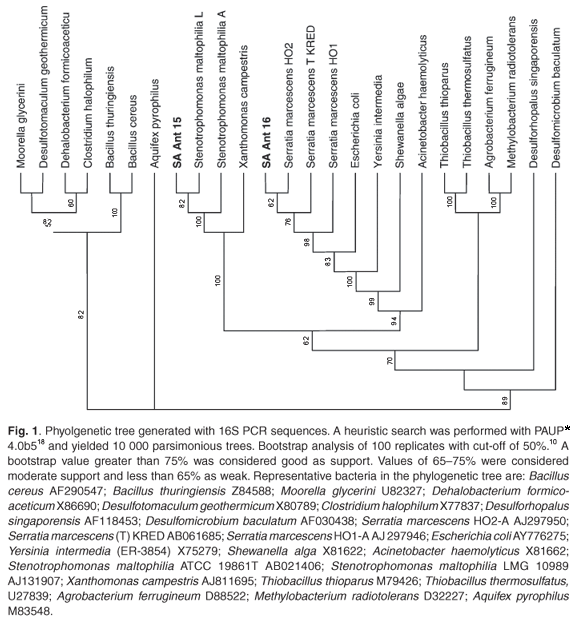
Two bacterial cultures, resistant to antimony and arsenate, were selected from the isolates. Near full-length 16S rDNA sequences were deposited in the NCBI database under accession numbers DQ 079059 and AY551938. BLAST (v 2.2.13) analysis, with entries available at the EMBL, GenBank, and Ribosomal Data Project (release 9.35) databases, retrieved an optimum alignment (98% identity) with the 16S rDNA of Serratia marcescens (AB061685) and (99% identity) with the 16S rDNA of Stenotrophomonas maltophilia (AJ131114), respectively. The phylogenetic tree (Fig. 1) illustrates the relatedness of these isolates to their counterparts. API and Biolog testing confirmed isolate SA Ant 16 as Serratia marcescens with a similarity index of 0.58. It was not possible to make a definitive identification using biochemical testing for isolate SA Ant 15.
Growth
The bacteria exhibited varying tolerance levels for both arsenite and arsenate. Stenotrophomonas maltophilia SA Ant 15 grew in up to 10 mmol l–1 arsenite and 20 mmol l–1 arsenate (see Fig. A in supplementary material online), while Serratia marcescens SA Ant 16 was able to grow in 15 mmol l–1 arsenite and in up to 500 mmol l–1 arsenate (Fig. B in online supplement). Both the specific growth rate and biomass yield were severely inhibited for S. maltophilia SA Ant 15 in the presence of arsenite. Arsenate inhibited growth less than arsenite, as indicated by specific growth rate and biomass yield, but longer lag phases were observed with the addition of increasing arsenic concentrations. For S. marcescens SA Ant 16, addition of arsenite resulted in a decrease in both specific growth rate and biomass up to a threshold concentration of 10 mmol l–1 with a sharp decline in both these parameters at higher concentrations. As for S. maltophilia SA Ant 15, addition of arsenate resulted in a proportionately lengthened lag phase. A linear decrease in total biomass yield was observed as well as a decline in specific growth rate up to 500 mmol l–1 arsenate. Results are summarized in Table 1.

Arsenate reduction by resting cells
Stenotrophomonas maltophilia SA Ant 15 removed 100% of the arsenate at a rate of 92.4 µmol l–1 h–1per mg cells over the first 2 h. Of all the arsenate removed, 25% of this removal rate could be attributed to reduction to arsenite at 4 µmol l–1h–1 per mg cells. Serratia marcescens SA Ant 16 removed 50% of the arsenate and, in total, 15% of the arsenate added was reduced to arsenite at approximately 2 µmol l–1h–1 per mg cells (Fig. 2). No chemical reduction was observed in control experiments.

Discussion
Two pure bacterial cultures, hyper-resistant to arsenate and arsenite, were isolated and identified as Stenotrophomonas maltophilia SA Ant 15 and Serratia marcescens SA Ant 16. The bacteria tolerated exceptionally high concentrations of arsenate (up to 500 mmol l–1 or ~37 500 ppm) and high concentrations of arsenite (up to 15 mmol l–1 or ~1125 ppm) during growth. A model bacterium such as E. coli has been shown to grow in up to 50 mmol l–1 arsenate,20 while bacteria isolated from arsenic-contaminated sites in New Zealand were unable to grow in arsenate concentrations exceeding 50 mmol l–1 (C.R. Anderson, pers. comm.). While arsenic concentrations in soils naturally vary from 0.1 ppm to 1000 ppm,21 at the collection site the range is between 10 ppm and 30 ppm. Arsenic becomes concentrated to up to 200 ppm during the refining process,22 which is still much lower than the tolerance level of the hyper-resistant bacteria isolated from samples collected from this site. Both the bacterial strains described in this study were resistant to these exceptionally high concentrations without prior exposure to such high levels of arsenic, suggesting that this trait is genetically intrinsic. This raises interesting questions as to the evolutionary processes and pressures that would sustain a characteristic such as this in an environment where it would seem redundant.
Biomass yield, growth rate and duration of lag phase depended on the type of oxyanion enrichment. Both isolates were more sensitive to arsenite and increasing concentrations of arsenate caused longer growth lag phases, lower biomass yields and lower growth rates. It is important to interpret these results in the context of a dynamic system where the combined effects of the oxyanion and the transformation products are biologically significant. Other factors, such as a change in external pH during growth,23 may also contribute.
Both bacteria were able to reduce arsenate and extrude the resulting arsenite, but a significant portion of arsenate removed (69–77%) was not recovered as arsenite. The differences between the removal and reduction rates indicate additional mechanisms of arsenate removal, such as adsorption of the negatively charged arsenic ions by oppositely charged amino groups in the bacterial cell walls,24,25 methylation following reduction of arsenate to arsenite,26 or sequestration by a range of cysteine-rich peptides such as γ-glutamylcysteine (γ-EC) and glutathione.26,27
The hyper-resistant bacteria described in this report thus have significant potential for bioremediation in environments with excess arsenic and its products.
This work was supported by a grant from the National Research Foundation.
1. Smedley P.L. and Kinniburgh D.G. (2002). A review of the source, behavior and distribution of arsenic in natural waters. Appl. Geochem. 17, 517–568. [ Links ]
2. Ahman D., Krumholtz L.R., Hemond H.F., Lovley D.R. and Morel F.M.M. (1997). Microbial mobilization of arsenic from sediments of the Aberjona Watershed. Environ. Sci. Technol. 31, 2923–2930. [ Links ]
3. Dowdle P.R., Laverman A.M. and Oremland R.S. (1996). Bacterial reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl. Environ. Microbiol. 62, 1664–1669. [ Links ]
4. Newman D.K., Beveridge T.J. and Morel F.M.M. (1997). Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63, 2022–2028. [ Links ]
5. Rittle K.A., Drever J.I. and Colberg P.J. (1995). Precipitation of arsenic during sulfate reduction. Geomicrobiol. J. 13, 1–12. [ Links ]
6. Cervantes C., Ji G., Ramirez J.L. and Silver S. (1994). Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 15, 355–367. [ Links ]
7. Turpein R., Pantsar-Kallio M., Haggblom M. and Kairesalo T. (1999). Influence of microbes on the mobilization, toxicity and biomethylation of arsenic in soil. Sci. Tot. Environ. 236, 173–180. [ Links ]
8. Say R., Yilmaz N. and Denizli A. (2003). Biosorption of cadmium, lead, mercury, and arsenic ions by the fungus Penicillium purpurogenum. Sep. Sci. Technol. 38, 2039–2053. [ Links ]
9. Stolz J.F. and Oremland R.S. (1999). Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23, 615–627. [ Links ]
10. Rosen B.P., Weigel U., Monticello R.A. and Edwards B.P.F. (1991). Molecular analysis of an anion pump: purification of the ArsC protein. Arch. Biochem. Biophys. 284, 381–385. [ Links ]
11. Ji G., Garber E.A.E., Armes L.G., Chen C-M., Fuchs J.A. and Silver S. (1994). Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry 33, 7294–7299. [ Links ]
12. Bennett M.S., Guan Z., Laurberg M. and Su X-D. (2001). Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl Acad. Sci. USA 98, 13577–13582. [ Links ]
13. Mukhopadhyay R., Shi J. and Rosen B.P. (2000). Purification and characterization of Acr2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 275, 21149–21157. [ Links ]
14. Lane D.J., Weisberg W.G., Barns S.M. and Pelletier D.A. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. [ Links ]
15. Thompson J.D., Gibson T.J. Plewniak F., Jeanmougin F. and Higgins D.G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 24, 4876–4882. [ Links ]
16. Altschul S.F., Gish W., Miller W., Myers E.W. and Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [ Links ]
17. Cole J.R., Chai B., Farris R.J., Wang Q., Kulam S.A., McGarrell D.M., Garrity G.M. and Tiedje J.M. (2005). The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucl. Acids Res. 33 (Database Issue), D294–D296. [ Links ]
18. Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and other Methods) Version 4. Sinauer Associates, Sunderland, MA. [ Links ]
19. Meerow A.W., Lehmiller D.J. and Clayton J.R. (2003). Phylogeny and biogeography of Crinum L. (Amaryllidaceae) inferred from nuclear and limited plastid non-coding DNA sequences. Bot. J. Linn. Soc. 141, 349–363 [ Links ]
20. Silver S., Budd K., Leahy K.M., Shaw W.V., Hammond D., Novick R.P., Willsky G.R., Malamy M.H. and Rosenberg H. (1981). Inducible plasmid-determined resistance to arsenate, arsenite, and antimony(III) in Escherichia coli and Staphylococcus aureus. J. Bact. 146, 983–996. [ Links ]
21. Cullen W.R. and Reimer K.J. (1989). Arsenic speciation in the environment. Chem. Rev. 89, 713–764. [ Links ]
22. Davis D.R., Paterson D.B. and Griffiths D.H.C. (1986). Antimony in South Africa. J. S. Afr. Inst. Min. Metall. 86, 173–193. [ Links ]
23. Anderson C.R. and Cook G.M. (2004). Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr. Microbiol. 48, 341–347. [ Links ]
24. Bai R.S. and Abraham T.E. (2003). Studies on chromium(VI) adsorption-desorption using immobilized fungal biomass. Bioresour. Technol. 87, 17–26. [ Links ]
25. Urrutia M.M. and Beveridge T.M. (1994). Formation of fine-grained metal and silicate precipitates on a bacterial surface (Bacillus subtilis). Chem. Geol. 116, 261–280. [ Links ]
26. Brunken H.S., Nehrhorn A. and Breunig H.J. (1996). Isolation and characterization of a new arsenic methylating bacterium from soil. Microbiol. Res. 151, 37–41. [ Links ]
27. Dhankher O.M., Li Y., Rosen B.P., Shi J., Salt D., Senecoff J.F., Sashti N.A. and Meagher R.B. (2002). Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and γ-glutamylcysteine synthetase expression. Nature Biotechnol. 20, 1140–1145. [ Links ]
This article is accompanied by supplementary material online (see www.sajs.co.za).
‡ Author for correspondence.E-mail: vheerde.sci@ufs.ac.za
Materials and methods
Enrichments
Soil and water samples were collected aseptically and placed on ice at the mining and refining sites of Consolidated Murchinson Mine near Gravelotte in the Limpopo province of South Africa. Because of the structural similarity between arsenic and antimony and the high concentrations of both at the site, aerobic enrichments were performed in TYG medium at pH 5.8, containing 5 g l–1 tryptone (Biolab), 3gl–1 yeast extract (Biolab),1gl–1 glucose (Holpro) and 100 mmol l–1 potassium antimony tartrate (Sigma) for two days at 37ºC. Positive enrichment cultures were transferred into fresh TYG medium supplemented with 100 mmol l–1 potassium antimony tartrate and incubated overnight at 37ºC and thereafter streaked on antimony-supplemented plates (100 mmol l–1) to obtain pure cultures. These pure cultures were then inoculated into two sets of media, consisting, respectively, of TYG media containing up to 250 mmol l–1 arsenate (Na2HAsO4) (BDH Chemicals) and TYG media enriched with up to 100 mmol l–1 arsenite (NaAsO2) (BDH Chemicals), and cultured at 37ºC in a rotary shaker at 200 rpm. Two isolates capable of growth in the presence of arsenic were used for further experiments.
Species identification
Genomic DNA was extracted with DNAZOLTM reagent (Gibco BRL), according to the manufacturer's instructions, and 16S rDNA fragments were amplified using universal bacterial primers 27F and 1492R.14 The PCR products were ligated into pGem®T-Easy vector (Pro-mega), transformed into competent E. coli JM109 cells, and plasmids containing inserts were isolated using standard procedures. Sequencing was performed on the 16S rDNA products from both isolates and aligned, using ClustalX (1.83),15 with those of bacteria previously found in the subsurface of mining environments as well as the closest matches revealed with BLAST searches16 and at RDP.17 A strict consensus tree18,19 was constructed and rooted with the outgroup Aquifex pyrophilus. GN2 MicroplatesTM (Biolog, Hayward) and API 20E panels (bioMerieux) were used to confirm the identification.
Minimum inhibitory growth concentration
Bacteria were inoculated into 30 ml TYG media in Erlenmeyer flasks at pH 5.8 and grown at 37ºC. TYG media, enriched with increasing concentrations of arsenite and arsenate, was inoculated with exponential growth phase cells to an OD560 of approximately 0.1. The inoculated cultures were grown with shaking at 37ºC, samples withdrawn hourly and optical density monitored at 560 nm over a 12-h period.
Whole cell arsenate reduction
Bacteria were grown at 37ºC in 100 ml TYG media containing 1 mmol l–1 arsenate until late exponential growth phase was reached. Cells were harvested by centrifugation at 8000 × g, washed in 10 mol l–1 PIPES buffer, pH 6.5 and resuspended in the same buffer (1:1 w/v). This was then supplemented with glucose (0.2% w/v) and arsenate (10 mmol l–1) and incubated at 37ºC. Aliquots were withdrawn, cells pelleted by centrifugation and the supernatant stored at –80ºC until analysis. A negative control, without any cells, was employed to monitor chemical reduction. Arsenic species were separated on a Hamilton PRP X-100 HPLC column (12 mmol l–1 H3PO4, pH 3.2 at 1 ml min–1, isocratic elution), and both substrate depletion (arsenate) and product formation (arsenite) were monitored at 195 nm.
Supplementary material to:
Botes E., Van Heerden E. and Litthauer D. (2007). Hyper-resistance to arsenic in bacteria isolated from an antimony mine in South Africa. S. Afr. J. Sci. 103, 279–281.
















